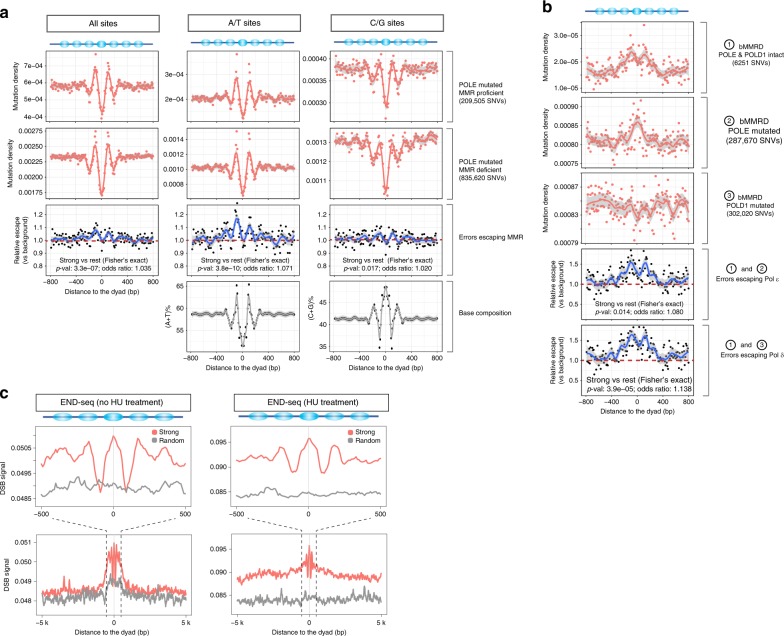
Fig. 4. Mismatch repair (MMR), DNA polymerase fidelity, and double-strand breaks (DSB) explain increased mutation rates in strong nucleosomes.
a Mutation density profiles relative to strong-nucleosome dyads in cancer genomes harboring driver mutations in the POLE and MMR pathway genes. Numbers of mutations used are indicated in the brackets. The MMR escape ratio compares the mutation densities in the MMR-proficient and MMR-deficient genomes. b Mutation density profiles relative to strong-nucleosome dyads for bMMRD cancer genomes with different driver mutation statuses in the POLE and POLD1 genes. The escape ratios compare the mutation densities for Pol ε-deficient and Pol δ-deficient cancers with the proficient ones. c END-seq signal indicating the density of DSBs relative to strong-nucleosome dyads. HU hydroxyurea. Two-sided Fisher’s exact test was used for testing the association of strong nucleosomal regions (dyad ± 95 bp) with differential MMR/polymerase performance. Source data are provided as a Source Data file.