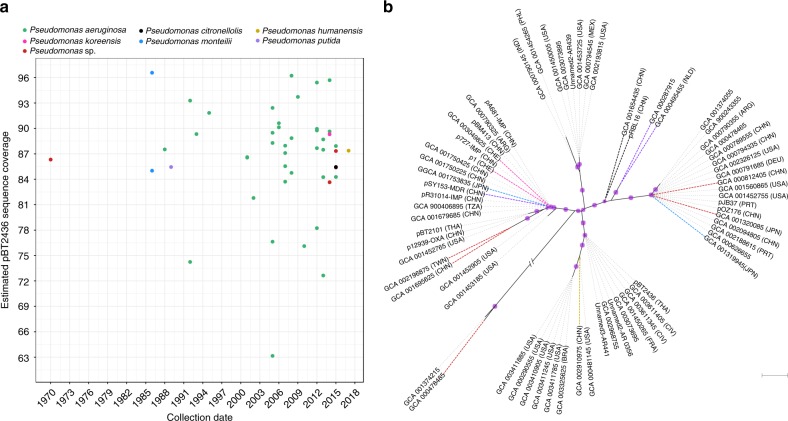
Fig. 6. pBT2436-like megaplasmids recovered from Pseudomonas genome assemblies deposited in GenBank.
Nucleotide sequences of Pseudomonas genomes from the four GenBank assembly categories were aligned to pBT2436 to identify unreported or overlooked related megaplasmids. Search was performed in November 2018. a The chart shows the pBT2436 estimated coverage and collection date reported in the metadata (when available) of matches covering >60% of the pBT2436 sequence (Supplementary Data 4). Bacterial species of the matches are colour coded and indicated in the figure. b Phylogeny of pBT2436-like megaplasmids recovered from Pseudomonas assemblies. The maximum-likelihood unrooted tree (GTR+F+ASC+R2 model) was inferred from the alignment of 4 core genes selected as phylogenetic markers (see “Methods”) and represents the relationships among 53 sequences from GenBank displaying >80% pBT2436 coverage and 15 complete megaplasmids (bold letters). Dotted lines connect branch tips with the corresponding taxa names and sources (three-letter codes in parentheses, where known). Lines corresponding to non-aeruginosa species’ sequences are colour coded as in a. Purple dots indicate posterior probability values >0.7. The scale bar corresponds to 0.1 expected substitutions per site. No clear correlations between cluster composition and geographical origin or bacterial species of its taxa are observed.