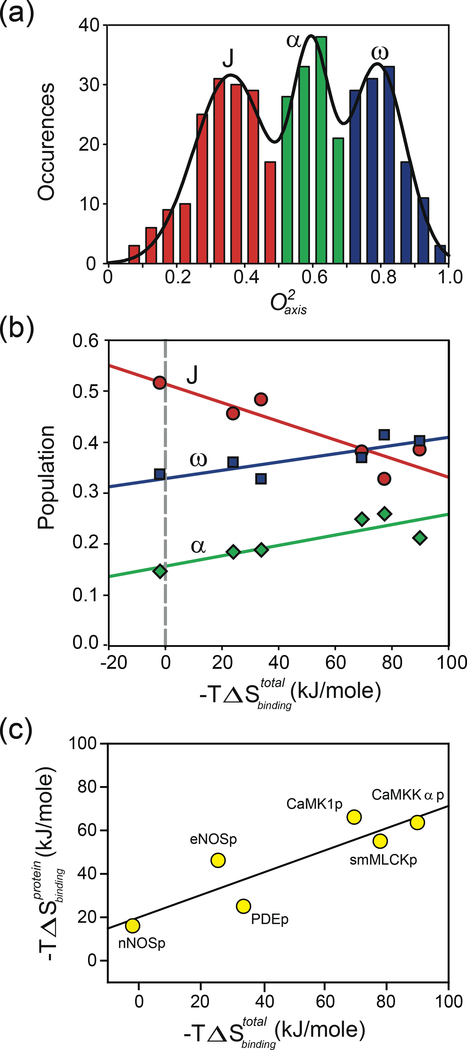
Figure 1.
Correlation of the apparent contribution of conformational entropy to the binding of target domains by calmodulin with total binding entropy. The apparent contribution reported by a model-dependent local interpretation of changes in methyl side-chain dynamics in calmodulin in complex with six different calmodulin-binding domains (19). (a) Summed histogram of the distribution of methyl group values of calmodulin in various complexes. (b) Correlation of the fractional population of the three classes of methyl-group motion with the total binding entropy measured by isothermal titration calorimetry for the six calmodulin complexes: J-class (red circles), α-class (green diamonds), and ω-class (blue squares). (c) Application of the oscillator inventory approach to assess the role of conformational entropy of calmodulin in the binding of calmodulin-binding domains. The harmonic oscillator potential was used to convert changes in methyl-group motion to estimates of changes in local side-chain entropy. The simple sum of apparent entropies is plotted against the total binding entropy. A The p at the end of the terms in panel c signifies that the various calmodulin-binding domains are represented by peptides. dapted from Reference 19. Abbreviations: CaMK1p, calmodulin-binding domain of calcium/calmodulin-dependent protein kinase1; CaMKKαp, calmodulin-binding domain of calcium/calmodulin-dependent protein kinase kinase alpha; eNOSp, calmodulin-binding domain of endothelial nitric oxide synthase; nNOSp, calmodulin-binding domain of neuronal nitric oxide synthase; PDEp, calmodulin-binding domain of phosphodiesterase; smMLCKp; calmodulin-binding domain of smooth muscle myosin light chain kinase.