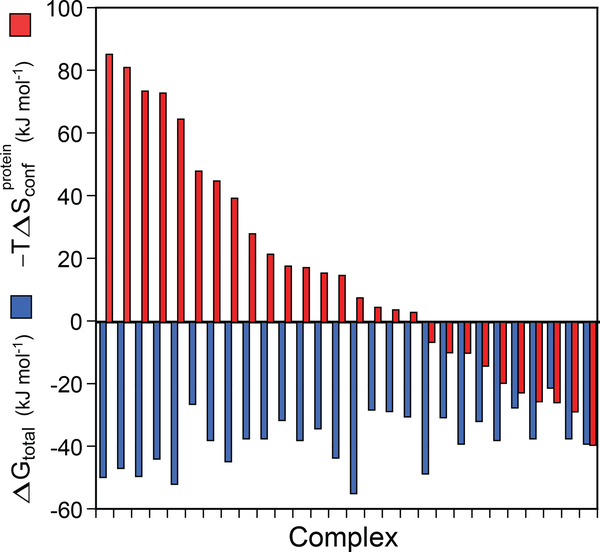
Figure 5.
Contribution of protein conformational entropy to the free energy of ligand binding to proteins. The broad range of contributions available to proteins for high-affinity binding of ligands is illustrated by the protein–ligand complexes used to calibrate the entropy meter (Figure 4). The 28 protein–ligand complexes are arranged in descending order of the contribution of conformational entropy (red bars) to the total free energy of binding (blue bars). Conformational entropy contributed by the response of amino acid side chains to the binding of a ligand can vary from highly unfavorable to negligible to highly favorable. In some cases, conformational entropy is essential for high-affinity binding. The structural origins of the variable utilization of conformational entropy in molecular recognition are unknown. In most cases, the change in solvent entropy remains a dominant contribution. Note that −TΔSr–t, ΔSligand, and ΔSsolvent are not shown here. Adapted from Reference 7