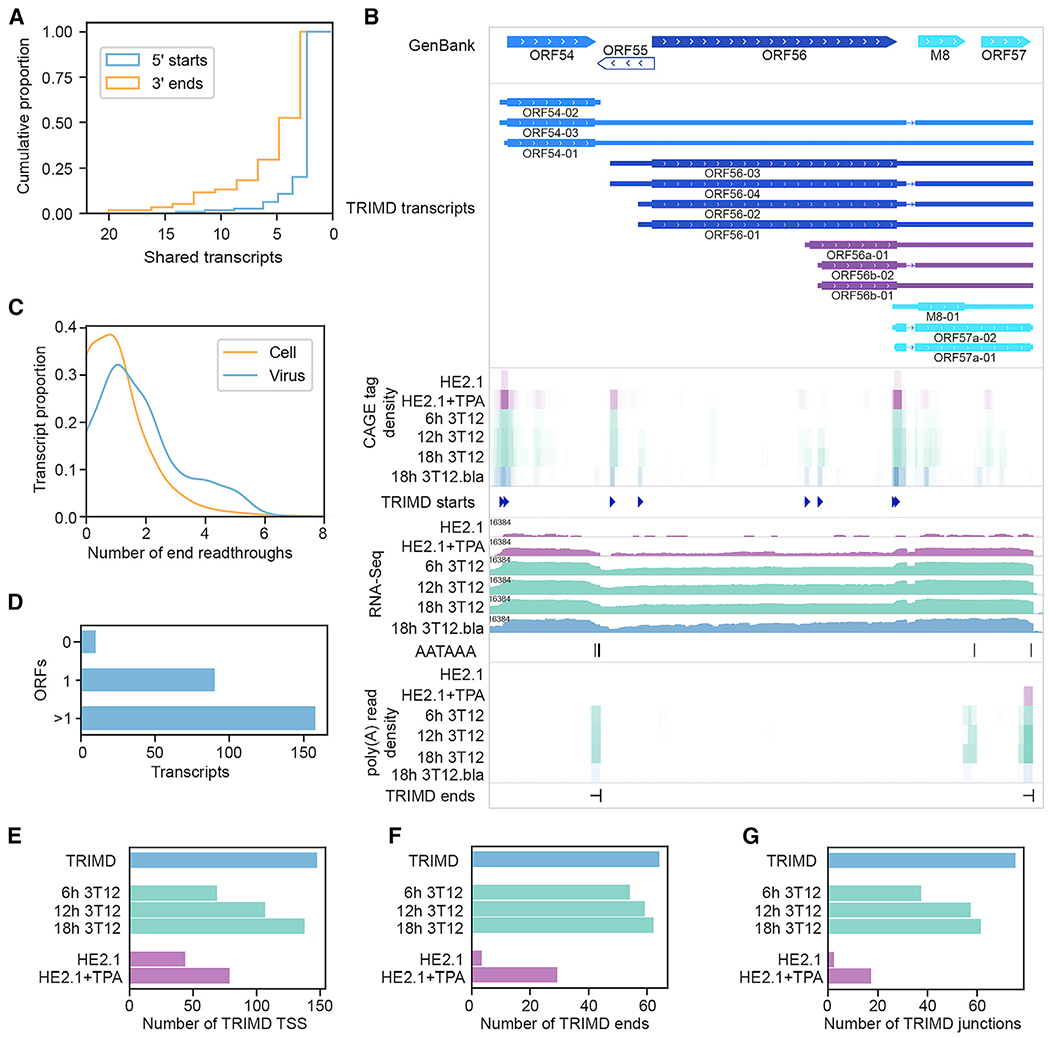
Figure 3. MHV68 Transcript Diversity.
(A) Cumulative proportion of viral 5′ TSSs and polyadenylation sites shared by ≥1 transcripts. One biological replicate, 3 sequencing methods.
(B) TRIMD-identified transcripts and supporting evidence at the ORF54-57 locus. The GenBank panel indicates reference GenBank-annotated ORFs in this locus. The TRIMD-identified transcripts are indicated (immediate early, bright blue; early, light blue; early-late, royal blue; late, dark blue; unknown ORF, purple), along with corresponding tracks for TRIMD-validated starts, poly(A) signal sequences, and ends. CAGE tag density, RNA-Seq, and poly(A) read density tracks are shown for TRIMD dataset (MHV68 marker virus infection of NIH 3T12 fibroblasts [3T12.bla]) plus validation sample sets (wild-type MHV68 infection of NIH 3T12 fibroblasts [3T12], latently infected HE2.1 B cells [HE2.1], and HE2.1 cells reactivated from latency [HE2.1 + TPA]). RNA-Seq read depth (log2 scale) is indicated. The M8/ORF57 splice variants ORF57a-01 and ORF57a-02 are not annotated in GenBank U97553, but they have been previously identified as variants of the immediate-early gene ORF57. Sequencing data tracks illustrate n = 1.
(C) Proportion of TRIMD-identified end readthroughs per transcript for cellular and viral transcripts. One biological replicate, 3 sequencing methods.
(D) Number of viral transcripts containing sequences corresponding to 0, 1, or >1 ORFs of ≥75 aa. One biological replicate, 3 sequencing methods.
(E) Number of TRIMD-identified viral TSSs detected at 6, 12, and 18 h following wild-type MHV68 infection of NIH 3T12 fibroblasts, in latently infected HE2.1 B cells, and 18h after the induction of HE2.1 B cell reactivation. The number of TRIMD-identified viral TSSs detected in TRIMD sample set (18h 3T12.bla) is shown for reference.
(F) Number of TRIMD-identified polyadenylation sites detected in validating datasets and TRIMD sample set indicated in (E).
(G) Number of TRIMD-identified splice junctions detected in validating datasets and TRIMD sample set indicated in (E).For (E)-(G) time course, and HE2.1 bars represent n = 1.