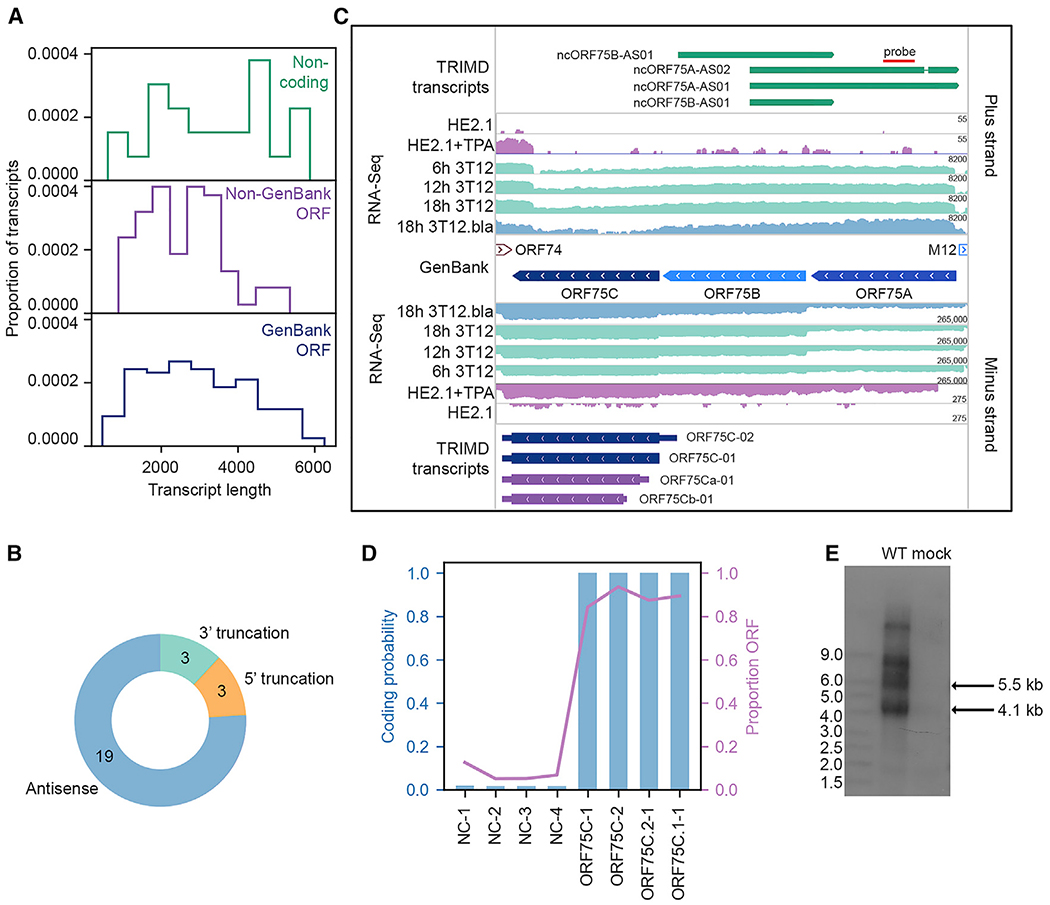
Figure 6. MHV68 Noncoding Transcripts.
(A) Proportion of noncoding transcripts, transcripts containing non-GenBank ORFs, and transcripts containing GenBank-annotated ORFs at specified transcript lengths.
(B) Type of noncoding transcript relative to overlapping ORFs.
(C) TRIMD-identified transcripts and supporting evidence at the ORF75A/B/C locus. The GenBank section indicates reference GenBank-annotated ORFs in this locus. TRIMD-identified transcripts are indicated (noncoding, dark green; early, light blue; early-late, royal blue; late, dark blue; unknown ORF, purple) for both plus and minus strands, along with corresponding RNA-seq tracks for the TRIMD dataset (18h 3T12.bla) and validation sample sets. RNA-seq read depth (log2 scale) is indicated. N = 1 for each sample. The red line indicates the probe site used in the northern blot in (E).
(D) CPAT-determined coding probability and ORF proportion for transcripts in the ORF75A/B/C locus.
(E) Northern blot for TRIMD-identified noncoding transcripts in theORF75 locus. NIH 3T12 fibroblasts were mock infected or were infected with wild-type MHV68 for 18 hours post-infection (hpi). The northern blot probe binding site is indicated in (C). Representative of 4 biological replicates.