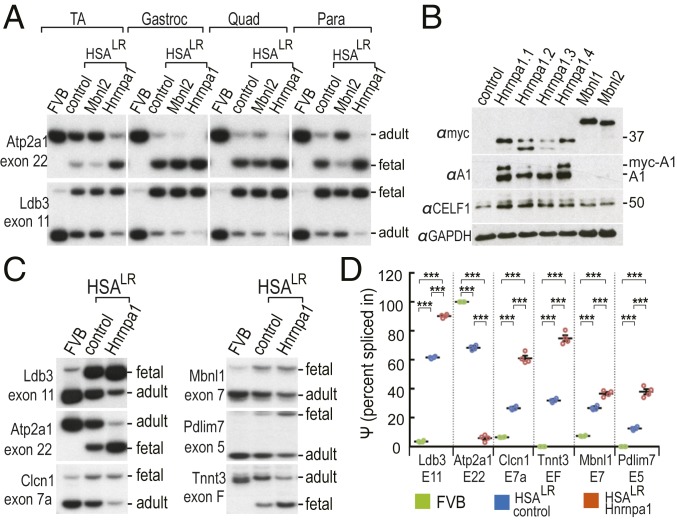
Fig. 2.
Increased expression of HNRNPA1 in skeletal muscles promotes fetal splicing patterns for DM1 targets. (A) Systemic HNRNPA1 overexpression exacerbated, while MBNL2 partially reversed, DM1 fetal splicing patterns in HSALR muscles. RT-PCR analysis of Atp2a1 exon 22 and Ldb3 exon 11 splicing patterns were used as representative DM1 targets and positions of the fetal and adult exon splicing RT-PCR products are indicated (Right). Alternative cassette exon splicing was determined in wild-type (FVB) and HSALR mice without injection (control) or following AAV9-Mbnl2 (Mbnl2) or AAV9-HnrnpA1 HSALR injections (n = 4 each). (B) Immunoblotting for myc-tagged HNRNPA1 (αmyc), HNRNPA1 (αA1), CELF1 (αCELF1), and the loading control GAPDH (αGAPDH) following direct i.m. injections of AAV9-mycHnrnpa1 (n = 4 mice Hnrnp1.1-1.4), AAV9-mycMbnl1, or AAV9-mycMbnl2. The positions of myc-HNRNPA1 (myc-A1) and endogenous HNRNPA1 (A1) are indicated. (C) RT-PCR splicing analysis of six DM1 targeted gene transcripts demonstrated that HNRNPA1 overexpression in HSALR TA promoted fetal splicing events. The wild-type splicing pattern for each gene is shown in the FVB lanes. (D) Fetal exon inclusion was determined using percentage spliced in (Ψ; n = 4). P values were calculated using a one-way ANOVA with Tukey’s HSD post hoc test. Data are SEM and significant. ***P < 0.001.