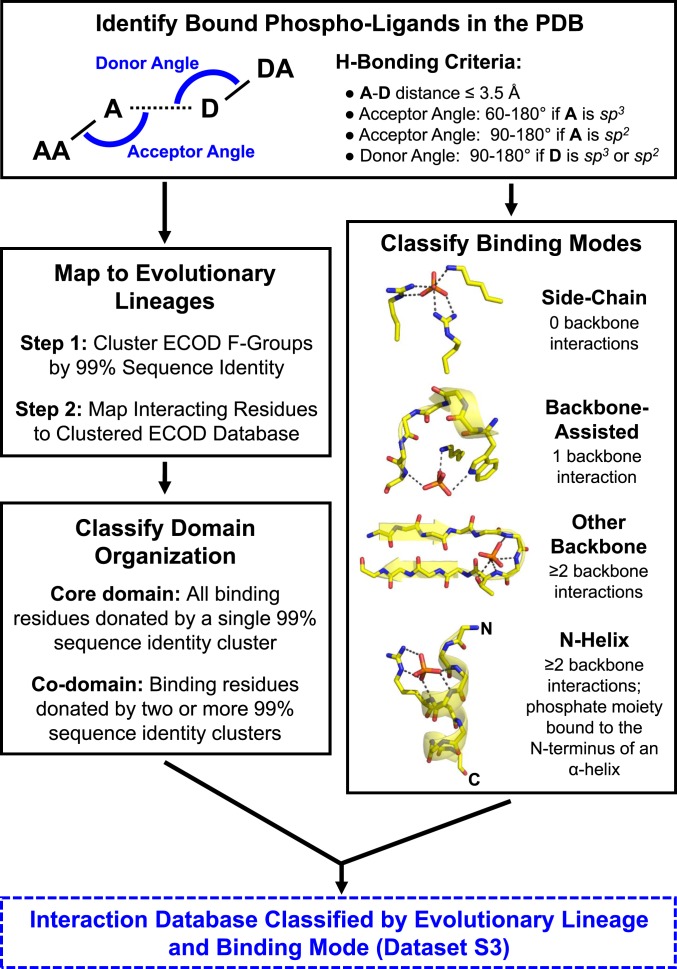
Fig. 1.
The phosphate-binding analysis pipeline (see Methods for additional details). Briefly, crystal structures in which any of the 100 most-common phospho-ligands, as well as phosphate and pyrophosphate, are bound were collected from the PDB (2). Interactions between the proteins and the phosphate moieties of these phospho-ligands were enumerated and used to classify the phosphate-binding mode. Four binding modes were defined, based on the extent that backbone amides participate in phosphate binding and whether the binding site is positioned at the N terminus of an α-helix. Interacting residues were also mapped to domains in the ECOD database. Subsequent analyses are therefore based on counting ECOD families (F-groups, which represent closely related structures) and independent evolutionary lineages (X-groups) rather than on individual PDB entries or binding events.