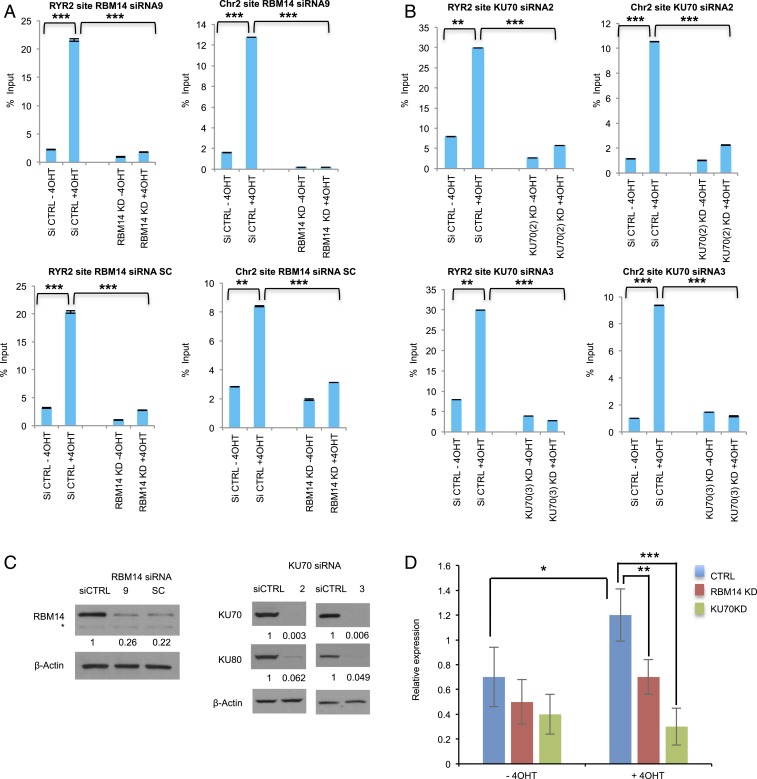
Fig. 4.
RNA:DNA hybrids are generated at DNA damage sites in a PARP-, transcription-, RBM14-, and KU-dependent manner. DRIP-qRT-PCR before I-PpoI activation (−4OHT) and after I-PpoI activation (+4OHT) at two individual I-PpoI sites, (RYR2 site, Chr2 intergenic site) in RBM14 (A) and KU70 (B) knockdown cells. The bar plot shows the percentage of enrichment relative to the input of total RNA:DNA hybrids with genomic DNA detected by primers near the I-PpoI sites. Data shown are representative of three independent experiments. Each experiment was performed in triplicate. **P < 0.01, ***P < 0.001 by two-tailed t test. (C) Knockdown levels of RBM14 and KU70 (for the DRIP experiments) are shown. The values indicated under the blot are the mean fold protein expression relative to control taken as 1 after normalization by β-actin (ImageJ quantification). siCTRL: control siRNA. *nonspecific band. (D) qRT-PCR analyses of nascent RNA expression at the Chr 2 site. Cells were transfected with I-PpoI and RBM14 and KU70 siRNA. Cells were collected, RNA was extracted, and qRT-PCR was performed at Chr 2 site. Nascent RNA was quantified using ∆∆Ct method. Cyclophilin B was used as a reference gene to calculate the ∆Ct value for each sample then each time point was normalized to the 0-h time point using ∆∆Ct formula. To calculate the fold change in gene expression, we took the 2 to the power of negative ∆∆Ct. Data shown are representative of three independent experiments. Each experiment was performed in triplicate.