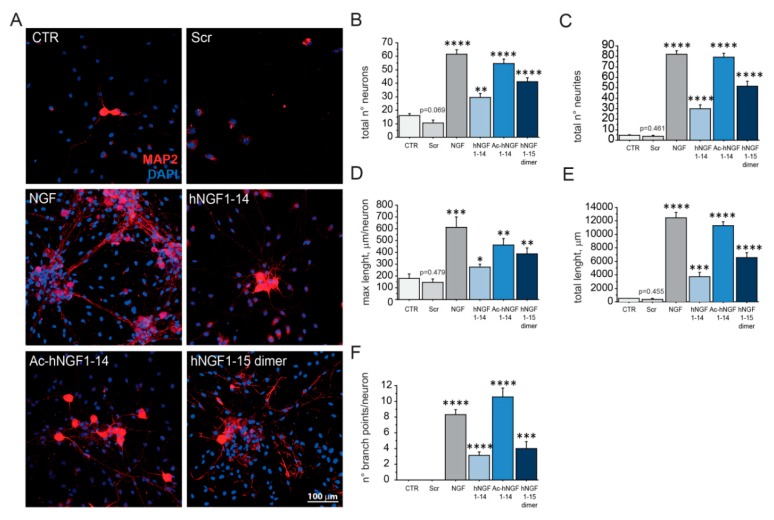
Figure 6.
hNGF1–14 peptides sustain survival and neurite elongation of DRG dissociated neurons in absence of NGF. (A-F) Classical DRG assay to test NGF-like bioactivity of hNGF1–14 peptides. (A) Dissociated DRG neurons plated and cultured for 5 days in presence of hNGF1–14 peptides (10 μM) or NGF (100 ng/mL; 3.84 nM). DRG neurons were fixed, immunolabelled with anti-MAP2 antibody for neurites staining, and nuclei were counterstained with DAPI. Images were acquired by means of confocal microscopy. Scrambled hNGF1–14 treated and untreated (CTR) DRG neurons survived although in lower number, and lacked of axons and neurites, typical of fully differentiated DRG neurons, as seen by absence of MAP2 staining (red colour). DRG neurons cultured with 10 μM hNGF1–14, 10 μM Ac-hNGF1–14, 10 μM hNGF1–15 dimer exhibit a mature DRG phenotype, including MAP2 positive neurite extension and an extensive neuronal network, resembling that promoted by the entire NGF molecule (NGF; 100 ng/mL; 3.84 nM, F). (B–F) DRG neurons survival and neurite outgrowth were analysed and quantified on MAP2 stained DIV5 DRG neurons by means of several parameters. (B) Number of surviving neurons: CTR (16.00 ± 1.58%), Scr (10.43 ± 2.30% of CTR; p = 0.069), NGF (122.06 ± 41.16% of CTR; **** p < 0.0001), hNGF1–14 (29.57 ± 2.85% of CTR; ** p < 0.01), Ac-hNGF1–14 (54.71 ± 3.41% of CTR; **** p < 0.0001), hNGF1–15 dimer (41.84 ± 2.94% of CTR; **** p < 0.0001). (C) Number of total neurites per field: CTR (4.57 ± 0.5%), Scr (3.71 ± 1.02; p = 0.461), NGF (82.14 ± 2.96; **** p < 0.0001), hNGF1–14 (29.86 ± 3.71; **** p < 0.0001), Ac-hNGF1–14 (79.00 ± 4.17; **** p < 0.0001), hNGF1–15 dimer (51.71 ± 4.71; **** p < 0.0001). (D) The maximum neurite length (length of the longest neuritis): CTR (180.87 ± 35.76 μm), Scr (146.67 ± 630.13 μm; p = 0.479), NGF (612.66 ± 87.60 μm; *** p < 0.001), hNGF1–14 (276.22 ± 23.87 μm; * p < 0.05), Ac-hNGF1–14 (460.92 ± 55.07 μm; ** p < 0.01), hNGF1–15 dimer (385.51 ± 52.61 μm; ** p < 0.01). (E) Total neurites length: CTR (479.02 ± 62.41 μm), Scr (383.86 ± 106.38 μm; p = 0.455), NGF (12471.32 ± 815.57 μm; **** p < 0.0001), hNGF1–14 (3753.74 ± 603.71 μm; *** p < 0.001), Ac-hNGF1–14 (11301.31 ± 548.43 μm; **** p < 0.0001), hNGF1–15 dimer (6586.87 ± 685.52 μm; **** p < 0.0001). (F) The number of total branch points: CTR (0), Scr (0), NGF (8.29 ± 0.64; **** p < 0.0001), hNGF1–14 (3.14 ± 0.40; **** p < 0.0001), Ac-hNGF1–14 (10.57 ± 1.13; **** p < 0.0001), hNGF1–15 dimer (4.00 ± 0.90; *** p < 0.001). Data (n = 7) were reported as mean +SEM. * p < 0.05; ** p < 0.01; *** p < 0.001, **** p < 0.0001. Scale bar = 100 μm.