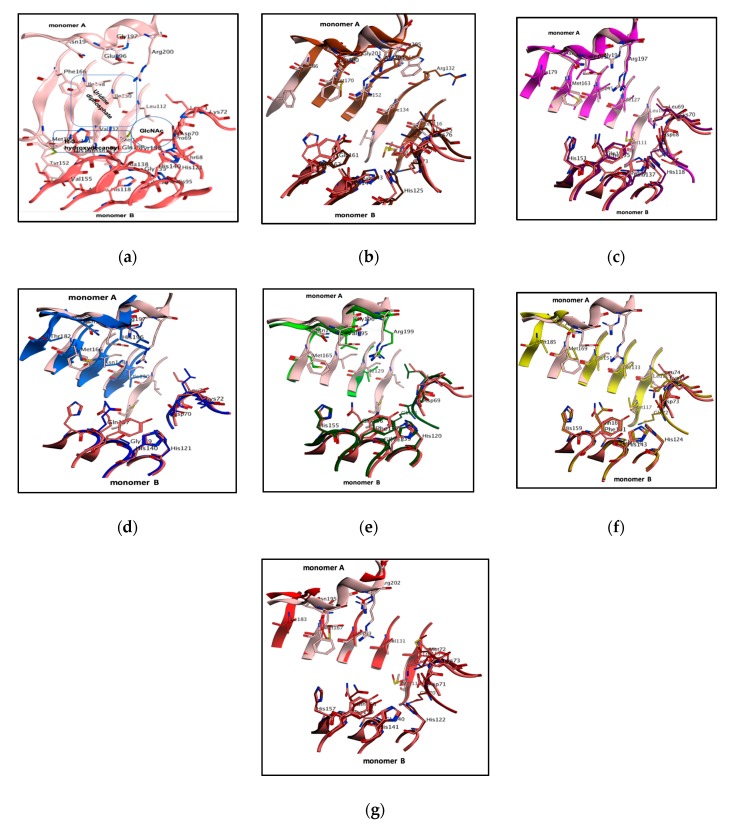
Figure 2.
The structural orientation of UDP-GlcNAc binding cavity and superimposition of PaLpxA with six different bacterial LpxAs (b-g) to design marked inhibitors. (a) The binding orientation of UDP-GlcNAc within the bifurcated PaLpxA active pocket, with regions highlighted as a square region for UDP binding, a circle for GlcNAc binding and a rectangular region for product binding. LpxAs are depicted as cartoons and colored with different orthologues, essential pocket residues are shown as classical sticks and colored with elements. Overlays of PaLpxA (light salmon for monomer A and dark salmon for monomer B) with (b) E. coli dimer (dark brown for monomer A and light brown for monomer B), (c) B. fragilis dimer (light magenta for monomer A and dark magenta for monomer B), (d) H. pylori dimer (light blue for monomer A and dark blue for monomer B), (e) L. interrogans dimer (light green for monomer A and dark green for monomer B), (f) A. baumannii (light yellow for monomer A and dark yellow for monomer B), and (g) B. thailandensis (light brown for monomer A and dark brown for monomer B).