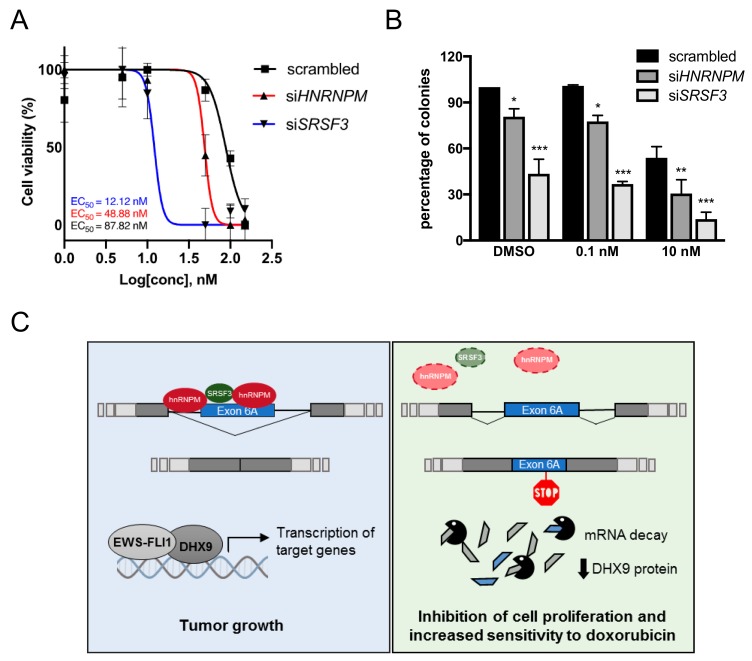
Figure 6.
Depletion of SRSF3 and hnRNPM increases doxorubicin sensitivity of Ewing sarcoma cells. (A) Dose-response curve of scrambled (black), siHNRNPM (red), and siSRSF3 (blue) TC-71 cells after treatment with increasing concentration of doxorubicin (from 0 to 150 nM). Cells were collected at 72 h after treatment. EC50 values are reported on the bottom. (B) Colony assay of scrambled, siSRSF3, and siHNRNPM TC-71 cells treated with increasing concentration of doxorubicin (from 0.1 nM to 10 nM in DMSO). Histogram shows the percentage of colonies determined 12 days after treatment. Statistical analysis was performed by two-way ANOVA. Asterisks indicate significance with Bonferroni post-hoc test (p-value: ***< 0.001, ** < 0.01, *< 0.05). (C) Graphical representation of the hypothetical regulatory mechanism driving DHX9 alternative splicing. On the left, hnRNPM and SRSF3 bind DHX9 pre-mRNA to induce the skipping of exon 6A. The translated full-length DHX9 protein can interact with EWS-FLI1 to promote transcription of target genes involved in cell proliferation and transformation. On the right, in the absence of either SRSF3 or hnRNPM, DHX9 pre-mRNA is processed to the exon 6a-included noncoding transcript targeted to NMD. The consequent reduction in DHX9 protein levels impacts Ewing sarcoma cells proliferation and sensitizes Ewing sarcoma cells to doxorubicin treatment.