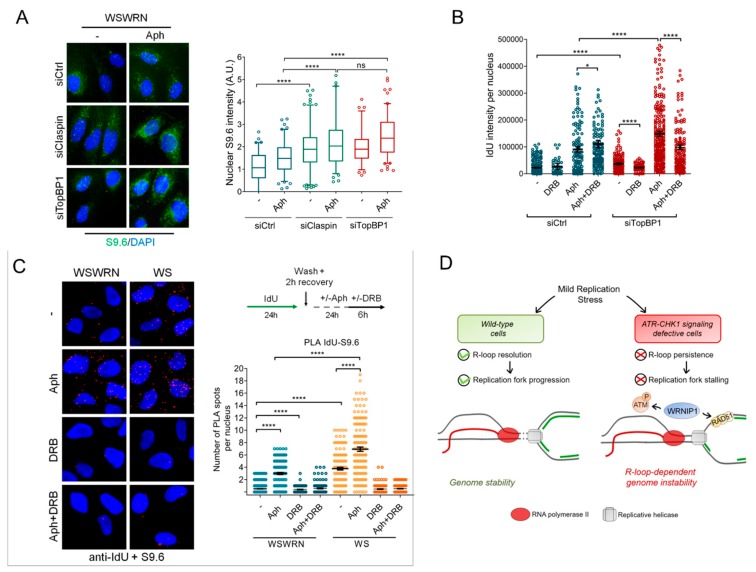
Figure 7.
Spatial proximity between ssDNA and R-loops upon MRS in cells with dysfunctional ATR-dependent checkpoint. (A) Immunofluorescence detection of R-loops with S9.6 antibody in WSWRN cells transfected with GFP, Claspin, or TopBP1 siRNA and treated with 0.4 µM Aph for 24 h. Box plot shows nuclear S9.6 fluorescence intensity. Box and whiskers represent 20–75 and 10–90 percentiles, respectively; (B) detection of ssDNA by IF with an anti-BrdU/IdU antibody in WSWRN transfected with GFP or TopBP1 siRNA and treated with 0.4 µM Aph for 24 h. Graph shows the mean ± SE from three independent experiments; (C) analysis of proximity between ssDNA (anti-BrdU/IdU antibody) and R-loops (anti-S9.6 antibody) performed by in situ PLA assay in WSWRN cells transfected with GFP or TopBP1 siRNA and treated as reported in the scheme. PLA interaction is shown in red. Graph shows the number of PLA spots per nucleus; and (D) proposed model of a role of WRNIP1 in suppressing R-loop-induced genome instability in cells with dysfunctional ATR-mediated checkpoint.