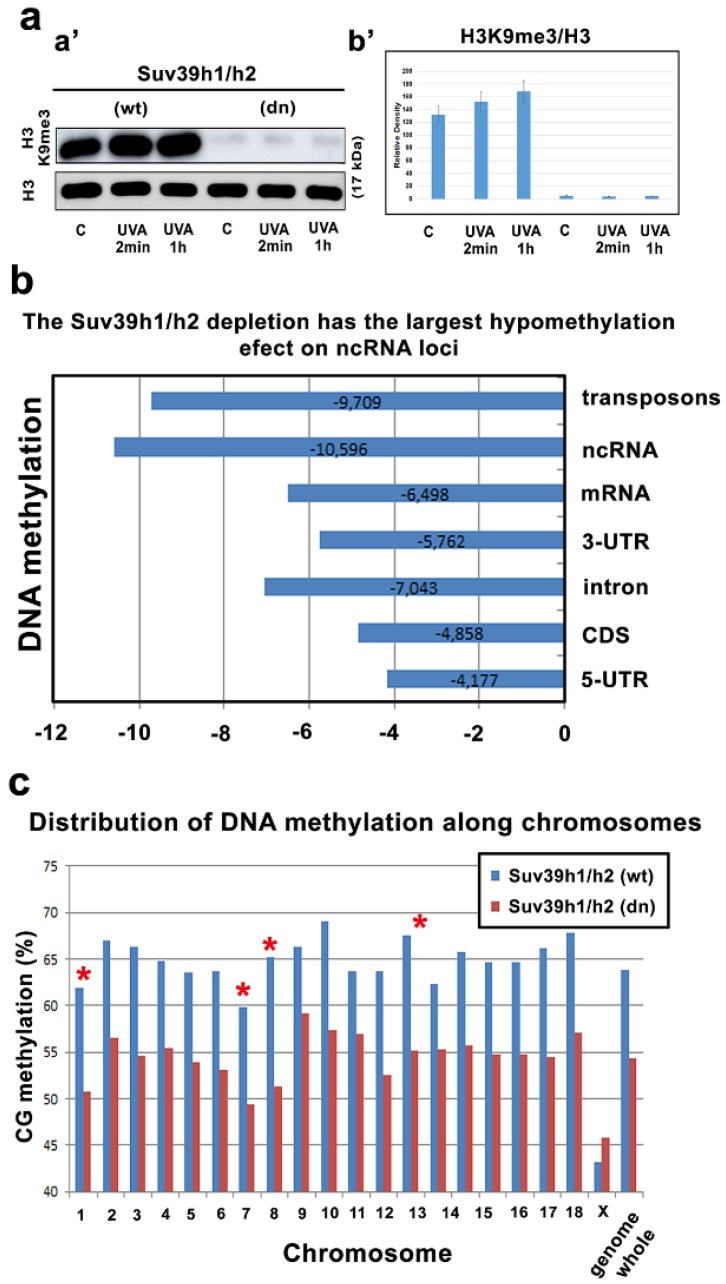
Figure 1.
Suv39h1/h2 depleted cells are characterized by global DNA hypomethylation. (a,a’) H3K9me3 is decreased in Suv39h1/h2 dn cells compared to the level in Suv39h1/h2 wt cells. (b’) Data from Western blot analysis were quantified by ImageJ software and normalized to the total protein levels and histone H3. (b) DNA methylation in distinct genomic regions was analyzed by whole-genome sequencing. The level of DNA methylation was studied in the Suv39h1/h2 wt and Suv39h1/h2 dn cells. (c) Global level of DNA methylation in individual autosomes and chromosome X in the Suv39h1/h2 wt and Suv39h1/h2 dn cells. Red stars show those chromosomes prone to DNA demethylation in Suv39h1/h2-depleted cells. Asterisks (*) show chromosomes characterized by the highest level of demethylation in Suv39h1/h2 depleted cells.