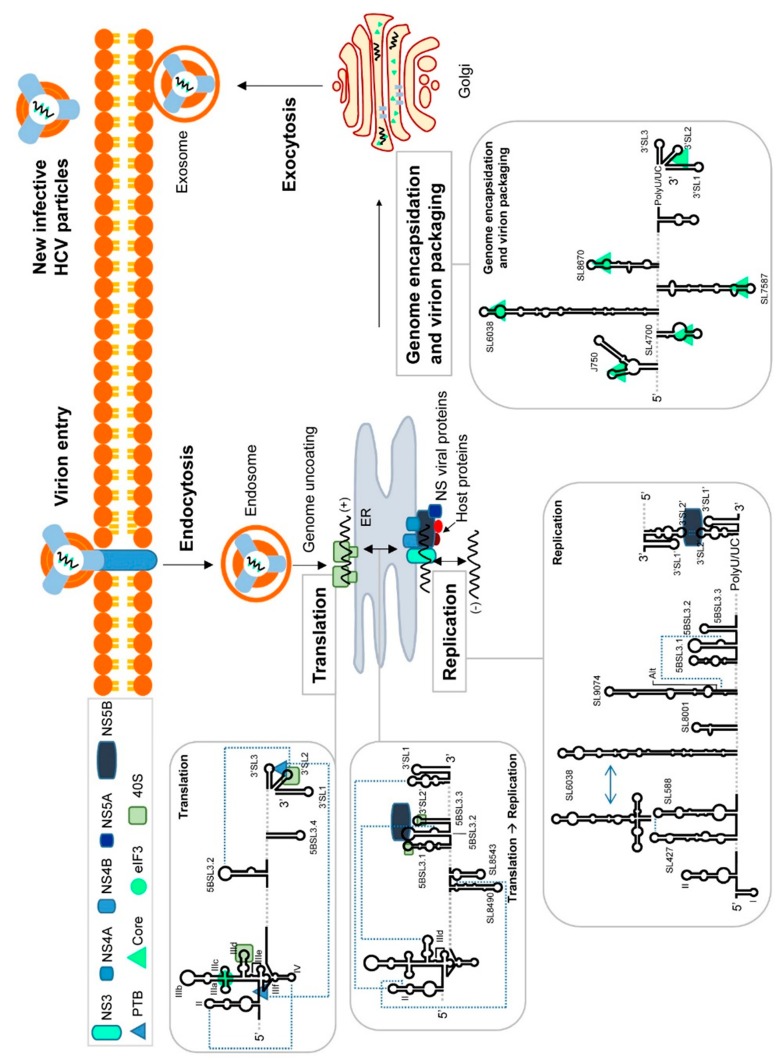
Figure 2.
Long-range RNA-RNA interactions and RNA structural domains control the HCV intracellular cycle. The figure shows a model of the viral cycle, outlining the participation of functional genomic domains and the RNA-RNA interactome in every step. Briefly, following the recognition of the viral envelope proteins by receptors on the hepatocyte surface, entry occurs by clathrin-mediated endocytosis. The endosome membrane and the viral envelope fuse and the capsid become disorganized (uncoating) - a process that requires the low pH of the endosome interior. The genomic RNA is directly translated on the surface of the endoplasmic reticulum (ER) in an IRES-dependent manner. IRES subdomain IIId is occupied by the 40S ribosomal subunit, impeding the interaction IIId-5BSL3.2, but favoring the contact 5BSL3.2-3′SL2. The interaction established between IRES domains II and IV provides an optimal structural environment for the correct positioning of the translation start codon. The translation is enhanced by the acquisition of a circular isoform mediated by the oligomerization of PTB which binds to the IRES and the 3′X tail. The synthesized polyprotein is co- and post-translationally processed to generate all the viral factors required for replication. Switching from translation to replication requires the formation of a closed-loop conformation mainly dependent on long-range RNA-RNA interactions: Domain II-SL8523; domain II-3′SL2′ and subdomain IIId-5BSL3.2. These contacts promote a translationally repressed-state and enhance replication via the recruitment of the NS5B viral polymerase and the 40S subunit to the functional region at the 3′ of the ORF. Viral RNA synthesis occurs in the replication complexes on the surface of the endoplasmic reticulum; this requires multiple RNA domains, but also important conformational rearrangements at the 3′X promoted by the interaction 5BSL3.2-Alt. This contact induces a structure at the 3′ end of the viral genome that is susceptible to forming genomic dimers, an optimum substrate for the NS5B protein. Finally, encapsidation requires multiple stem-loops that work in a cooperative manner to ensure the packaging of full-length, intact viral RNA genomes. This process occurs in the ER and in the Golgi apparatus, yielding mature virions that are released to the extracellular medium by conventional exocytosis.