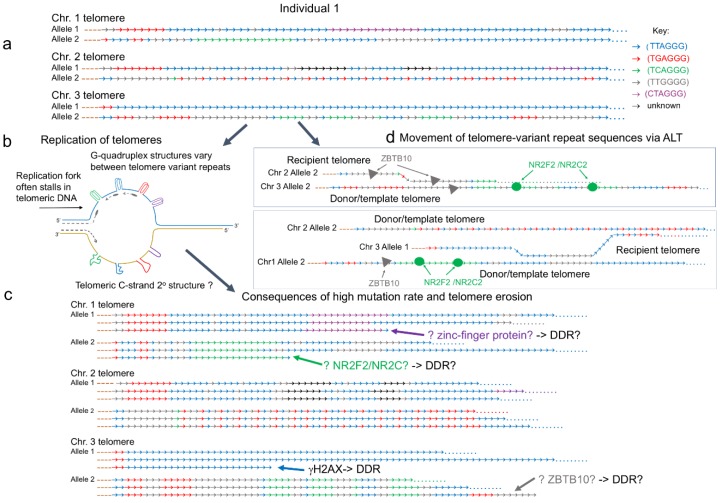
Figure 1.
Mutation processes at telomeres. Examples of the interspersion patterns of the canonical (TTAGGG) and telomere-variant repeats (TVRs) commonly found at the proximal (centromeric) ends of human telomeres. (a) The diagram represents three telomeres on different chromosomes (Chr. 1 to 3) each with different interspersion patterns in the two alleles. While many telomeres comprise a region of degenerate repeats at the proximal end, some telomeres lack sequence variation in this region, as shown in Chr. 3 allele 1. (b) During S phase, the replication fork is prone to pausing or stalling as it passes through telomeric DNA. This is due to secondary structures, such as t-loops (not shown) that must be unwound. In addition, a variety of G-quadruplex structures can form on the G-rich strand. It is not known if the C-strand can also adopt secondary structures that impede replication. These obstacles contribute to high somatic and germline mutation rates that usually result in gains or losses of repeats. (c) Telomere lengthening by the break-induced-replication (BIR) mechanism that underlies the alternative lengthening of telomeres (ALT) moves TVRs between telomeres and can result in the distribution of the variant repeats along the full length of the telomeric DNA. The NR2F2 and NR2C2 orphan nuclear receptors can bind to (TCAGGG)n containing telomeres in ALT positive cells. Similarly, ZBTB10 has recently been shown to bind to (TTGGGG)n repeats in ALT positive cells. The binding of these proteins at telomeres may contribute to an altered DNA damage response. (d) The replication-driven high mutation rate at telomeres creates mosaicism within normal tissues, cancers, and the germline such that some cells carry mutated telomere interspersion patterns. In addition, the replication-dependent erosion of telomere length results in short telomeres that trigger a DNA damage response (DDR). Telomere erosion may lead to exposure of sequence-variant repeats at the end of telomeric DNA. It is not known whether this triggers specific proteins to bind to the short telomeres, for example NR2F2, NR2C2, ZBTB10, or other zinc-finger containing proteins that may modify the DDR.