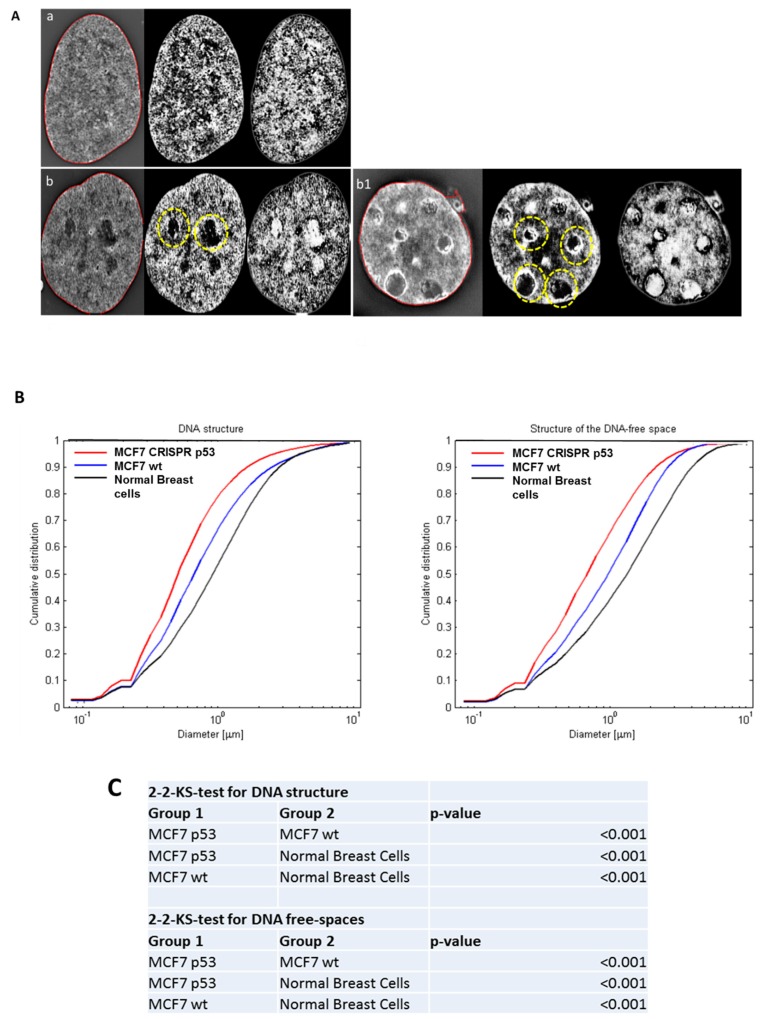
Figure 1.
The use of three-dimensional (3D)-SIM (Structured Illumination Microscopy) to investigate differences in DNA structure between normal breast cells, and MCF-7 (Michigan Cancer Foundation-7). (A) 3D-SIM images of primary normal breast cells (a), MCF7 wild type (b) and MCF7 CRISPR(Clustered Regularly Interspaced Short Palindromic Repeats) p53 deleted (b1). MCF7 shows more DNA-poor spaces than primary breast cells and CRISPR p53 deleted cell lines shows more DNA-poor spaces than the wild-type ones. Left panels: reconstructed 3D-SIM images; middle panels: light granulometry images; right panels: dark granulometry images. (B) Comparisons of granulometry DNA structure and DNA-poor spaces between the three cell lines. (C) p-value comparisons for the comparisons in (B).