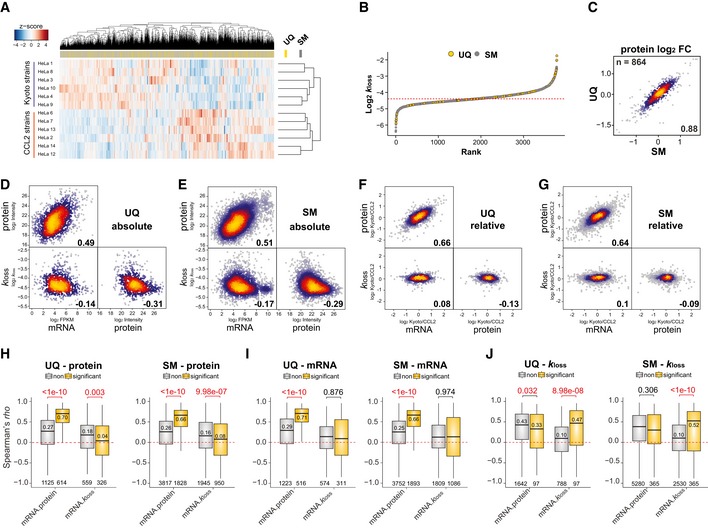
-
A
Hierarchical clustering of the k
loss values quantified for twelve HeLa cell lines. The positions of unique (UQ) and shared major (SM) proteins were highlighted in the separated bar.
-
B
Rank distribution of the averaged k
loss values of UQ and SM proteins across HeLa cells. The dashed red line indicates median.
-
C
Correlation of Kyoto/CCL2 protein fold change for IDs quantified in both UQ and SM.
-
D, E
Across‐gene Spearman's absolute correlation between indicated values (average from all cell lines).
-
F, G
Spearman's correlation between indicated values using relative quantification data (i.e., Kyoto/CCL2 fold change).
-
H–J
Protein AS group‐specific Spearman's rho distribution of isoforms that were differentially expressed on mRNA level (I, EdgeR, adjusted P < 0.01), protein level (H, t‐test, adjusted P < 0.01), or degraded (J, t‐test, adjusted P < 0.01) between Kyoto and CCL2 (yellow) and which are not (gray). The numbers indicate Wilcoxon P‐values (top), median rho of significant comparisons (above or below the median bar), and number of values (bottom). Spearman's rho was calculated for every protein AS group in the data set using twelve data points and summarized across genes/proteins. Box borders represent the 25th and 75th percentiles, bar within the box represents the median, and whiskers represent the minimum and maximum value within 1.5 times of interquartile range. The dashed red line indicates zero.