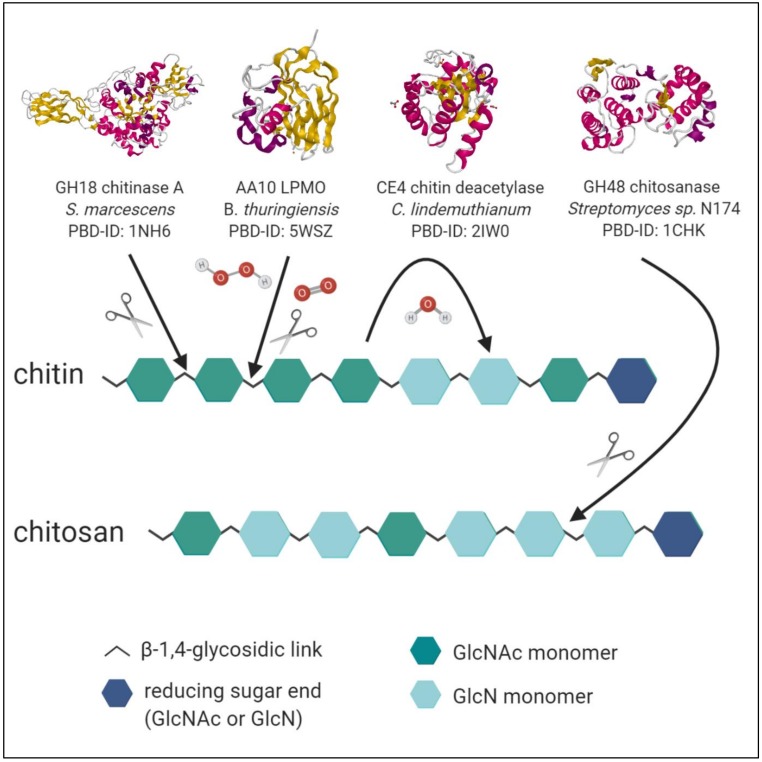
Figure 3.
Enzymes with catalytic activity towards chitin and chitosan. The crystal structures of the chitinase A from Serratia marcescens [95], the lytic polysaccharide monooxygenase (LPMO) from Bacillus thuringiensis [96], the chitin deacetylase (CDA) from Colletotrichum lindemuthianum [97] and the chitosanase from Streptomyces sp. N174 [77], derived from the RCSB protein data bank (PDB), are illustrated as exemplary enzymes of their respective catalytic activity. Red coloured parts are α-helices, while β-sheets are indicated by yellow. Chitinases generally hydrolyse the β-(1,4)-glycosidic links between two GlcNAc monomers. Their modes of action, amino sequence, and catalytic sites can vary substantially. LPMOs are copper-dependent enzymes, which cleave chitin by oxidation of C1 or C4. They can recruit either H2O2 or O2 as co-substrate and need an external reducing agent. The NodB-related CDAs hydrolyse the acetamido groups of GlcNAc monomers with a catalytic water molecule. Each enzyme exhibits specificity for target substrate sequences and creates unique deacetylation patterns. Chitosanases generally hydrolyse the β-(1,4)-glycosidic links between two GlcN monomers with a retaining or inverting mechanism. The specificity towards an additional substrate besides GlcN-GlcN is common. The figure was created with BioRender.