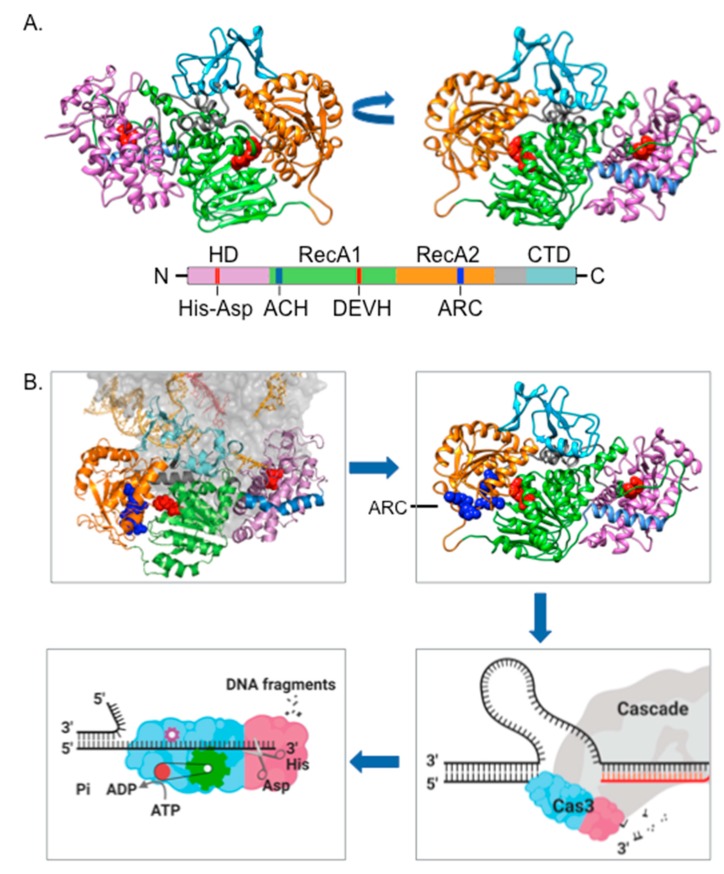
Figure 2.
Cas3 structure-function. A. PHYRE2-predicted Escherichia coli Cas3 structure, modeled from Cas3 structures solved from Thermobaculum terrenum (PDB 4Q2C) and Thermobifida fusca (PDB 4QQW, 4QQX and 4QQY), represented in two orientations and with a corresponding cartoon of primary sequence presented below. We highlight the HD domain (purple), two RecA-like domains (green and orange) and the accessory C-terminal domain (CTD, pale blue). Active sites comprising the Asp-His HD domain and the amino acid DEVH motif of one RecA-like domain for ATP-hydrolysis are highlighted in red spheres within the structures, and marked on the cartoon primary structure. Also marked are the prominent solvent-exposed alpha helix (ACH) and an arginine rich channel (ARC) described in the main text. B. Panels should be followed from top left, clockwise. The location of Cas3 (using same domain colors as used in part A) bound to Cascade subunit protein Cse1 (grey), presented from the T. fusca Cascade-Cas3 structure (PDB: 6C66). DNA parts of the R-loop are colored orange—in CRISPR interference this DNA is nicked by the Cas3 HD domain, and then assimilated into the translocase/helicase active sites, possibly via interaction between DNA and the arginine residues of the ‘ARC’, highlighted as blue spheres. Captured single-strand DNA (ssDNA) is then translocated through the Cas3 protein by a reeling mechanism, which is associated with nuclease activity that generates DNA fragments.