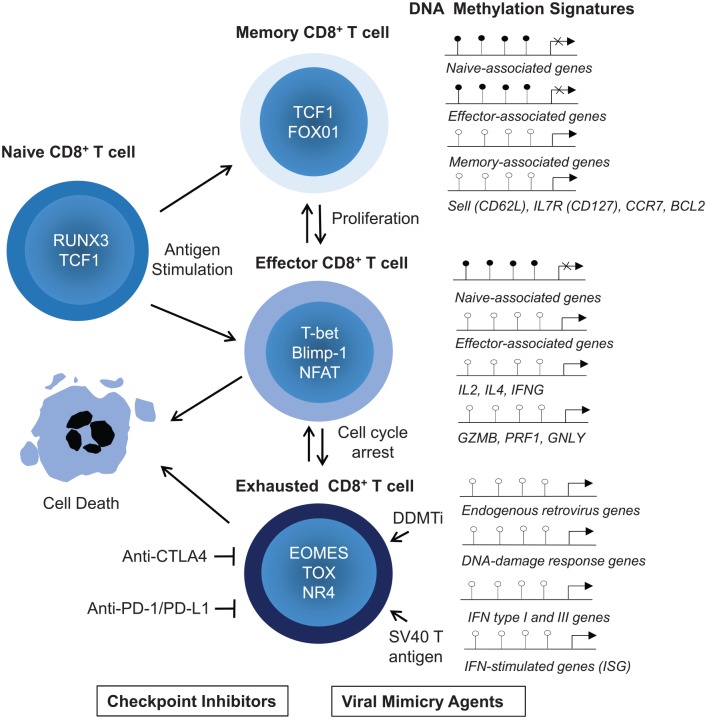
Figure 2.
Genetic and epigenetic regulators of CD8+ T-cell activation and differentiation into memory, effector, and exhausted T-cell populations. DNA demethylation at CpG sites of regulatory regions and locus and action of master transcriptional factors (TCF1, T-bet) result in the transcriptional repression or derepression of key genes associated with memory (sell, CCR7, IL7R), survival (BCL-2, CD127), proliferation (IL-2, IL-4), and effector functions (IFNs, GZMB, PRF1, KLRG1, GNLY). Effector T cells die upon antigen withdrawal or overactivation. DDMTi and SV40 large T antigen promote viral mimicry state and derepression of human endogenous retrovirus (HERVs) that activate the MDA5/RIG/MAVS innate immune response pathways. This leads to type I and III interferon-mediated synthesis of IGSs. A combination of viral mimicry agents and inhibitory antibodies to immune checkpoint molecules, such as CTLA-4, PD-1, PDL-1, and PDL-2, may reverse CD8+ T cell-exhaustion through epigenetic reprogramming.
Blimp-1, B-lymphocyte-induced maturation protein-1; DDMTi, DNA demethylating agents; GNLY, granulysin; GZMB, granzyme B; IFN, interferon; IGSs, interferon-stimulated genes; IL, interleukin; IL7R, interleukin 7 receptor; MAVS, mitochondrial antiviral-signaling protein; MDA5, melanoma differentiation-associated protein; PRF1, perforin; RIG I, retinoic acid-inducible gene I protein; RUNX, the Run-related domain for DNA binding and core-binding factor β complex; TCF1/Tcf-7, T-cell-specific high Mobility Group Box protein transcription factor 7; TOX, Thymocyte Selection Associated High Mobility Group Box protein; l to o = methylated (pin head closed in black), o = unmethylated (pin head opened in white).