Table 3.
Gas chromatography–mass spectrometry (GC-MS) analysis of the cypermethrin degradation metabolites.
| Cypermethrin Degradation Metabolites Sequence | Retention Time (min) | Identified Metabolites | Molecular Weight (MW) | Chemical Structure |
|---|---|---|---|---|
| CP1 | 4.123 | Phenol | 94 |
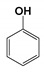
|
| CP2 | 11.050 | Benzoic acid, 2,5-dimethyl | 150.17 |
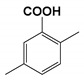
|
| CP3 | 13.724 | 2-Hydroxy-3- phenoxy- benzeneacetonitrile | 225 |
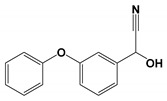
|
| CP4 | 13.728 | 3-Phenoxybenzaldehyde | 198 |
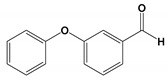
|
| CP5 | 14.289 | Phthalic acid | 166.14 |
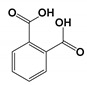
|
| CP6 | 15.942 | 2-Pentadecanone | 226.4 |
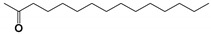
|
| CP7 | 20.00 | 3-Phenoxybenzoate | 228 |
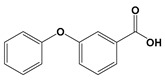
|
| CP8 | 23.960 | Cypermethrin | 415 |
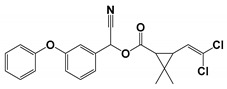
|
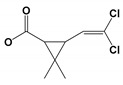
| CP9 | 24.098 | 3-(2,2-Dichloroethenyl)-2,2-dimethyl cyclopropanecarboxylate | 236 |
|
