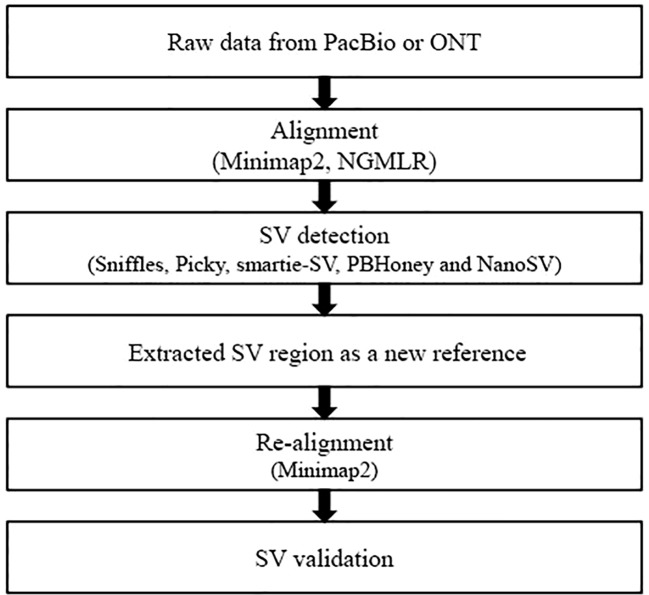
Figure 1.
Overview of the evaluation pipeline in this study. Minimap2 (Li, 2018) and NGMLR (Sedlazeck et al., 2018) were used to perform alignment. Minimap2 aligned against reference genome with parameters ‘–MD -x map-pb/map-ont -R “@RG\tID:default\tSM : SAM” -a’ and NGMLR with the default parameters. SVs were identified by five callers, including Sniffles (Sedlazeck et al., 2018), Picky (Gong et al., 2018), smartie-sv (Kronenberg et al., 2018), PBHoney (English et al., 2014), and NanoSV (Cretu Stancu et al., 2017). Sniffles detected all types of SVs with parameters ‘–genotype –skip_parameter_estimation –min_support 10’ and employed a novel SV scoring scheme to exclude false SVs based on the size, position, type, and coverage of the candidate SVs (Sedlazeck et al., 2018). NanoSV was set ‘-s samtools’ to detect SVs and used the clustering of split reads to identify SV breakpoint junctions based on long-read sequencing data (Cretu Stancu et al., 2017). PBHoney considered both intra-read discordance and soft-clipped tails of long read (>10, 000 bp) to identify SVs (English et al., 2014). PBHoney, smartie-sv, and Picky were used to identify SVs with default parameters.