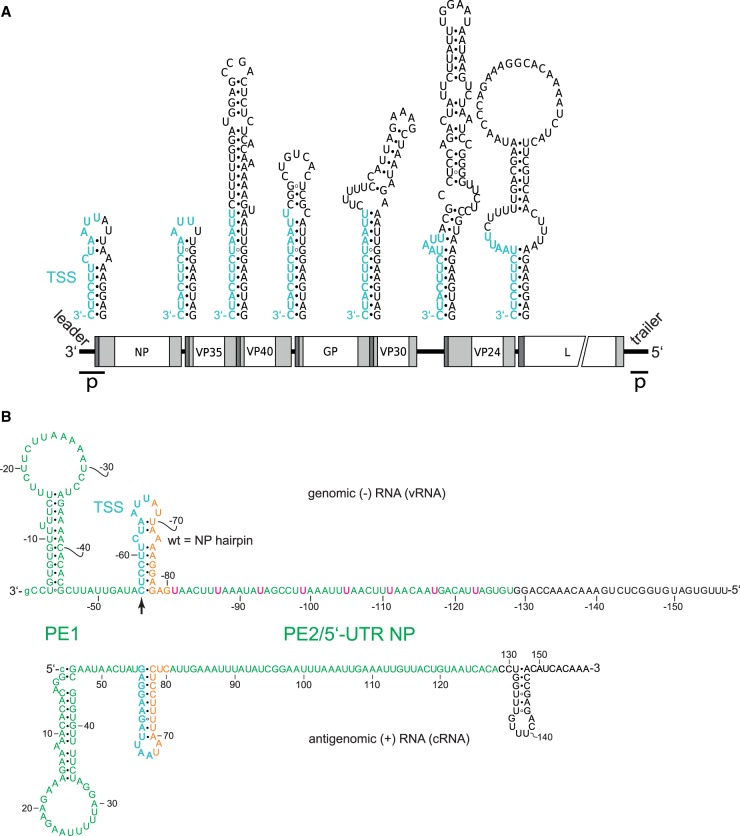
FIGURE 1.
Secondary structure formation potential at (A) transcription start regions of EBOV genes and (B) specifically in the genomic 3′-leader and the complementary antigenomic RNA. (A) Genomic sequence elements required for transcription (re)initiation are shown in cyan at the top; transcription is initiated opposite to the 3′-terminal C residue of the TSS. Schematic white boxes at the bottom mark the reading frames for EBOV proteins NP, VP35, VP40, GP, VP30, VP24 and L; light gray boxes indicate 5′- and 3′-UTRs, with dark gray areas depicting the position of the predicted secondary structures illustrated above the genome; p, location of leader and trailer promoters (Calain et al. 1999). (B) Validated secondary structures forming in the naked genomic leader RNA (top) and its complementary antigenomic RNA (bottom) (Weik et al. 2002, 2005; Schlereth et al. 2016); in the text and following figures, the genomic RNA is abbreviated as vRNA for viral RNA, and the antigenomic RNA as cRNA for complementary or copy RNA; the numbering of nucleotides in the genomic RNA strand is indicated by a minus sign preceding the nucleotide number. Proposed promoter elements 1 and 2 (PE1, PE2) are shown in green letters and the 3′-U residues of the eight UN5 hexamers in PE2 are highlighted in pink in the genomic RNA; orange nucleotides mark the spacer sequence between the transcription start sequence (TSS, in cyan) and PE2. The transcription initiation site on the genomic RNA is marked by the vertical arrow. Nucleotide numbering of the genomic RNA starts at the 3′-terminal nt (position −1) that is complementary to position 1 (5′-terminus) of the antigenome. The 3′-terminal G of the genome is shown as small letter to consider the recent finding that the presence of this nucleotide is not essential as the EBOV RNA polymerase initiates polymerization at the C residue preceding the 3′-terminal G residue (Deflubé et al. 2019).