Fig. 1.
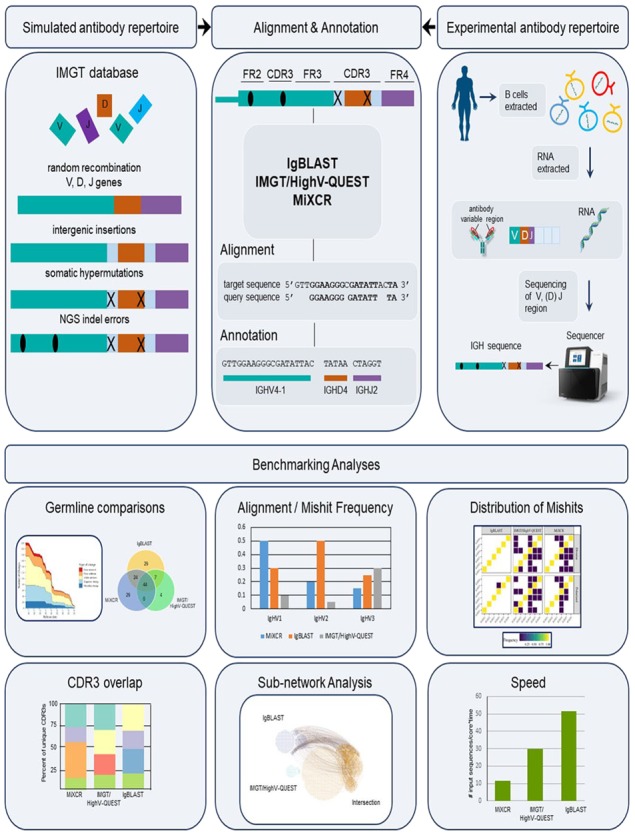
Schematic of immuneoinformatic benchmarking analysis. Top, Left side: Antibody repertoire data were simulated in silico using IgSimulator. Right side: Experimental antibody repertoire data was collected from publically available datasets. Top, Middle: Alignment and annotation of simulated and experimental sequences were performed with the immunoinformatic tools: IgBLAST, IMGT/HighV-QUEST and MiXCR. Bottom: Downstream analysis of the results compared: (i) the germline databases (ii) alignment (iii) frequency of mishits of the V, D and J gene annotations (iv) unique CDR3 overlap (v) sub-network analysis and (vi) speed of analysis
