Fig. 3.
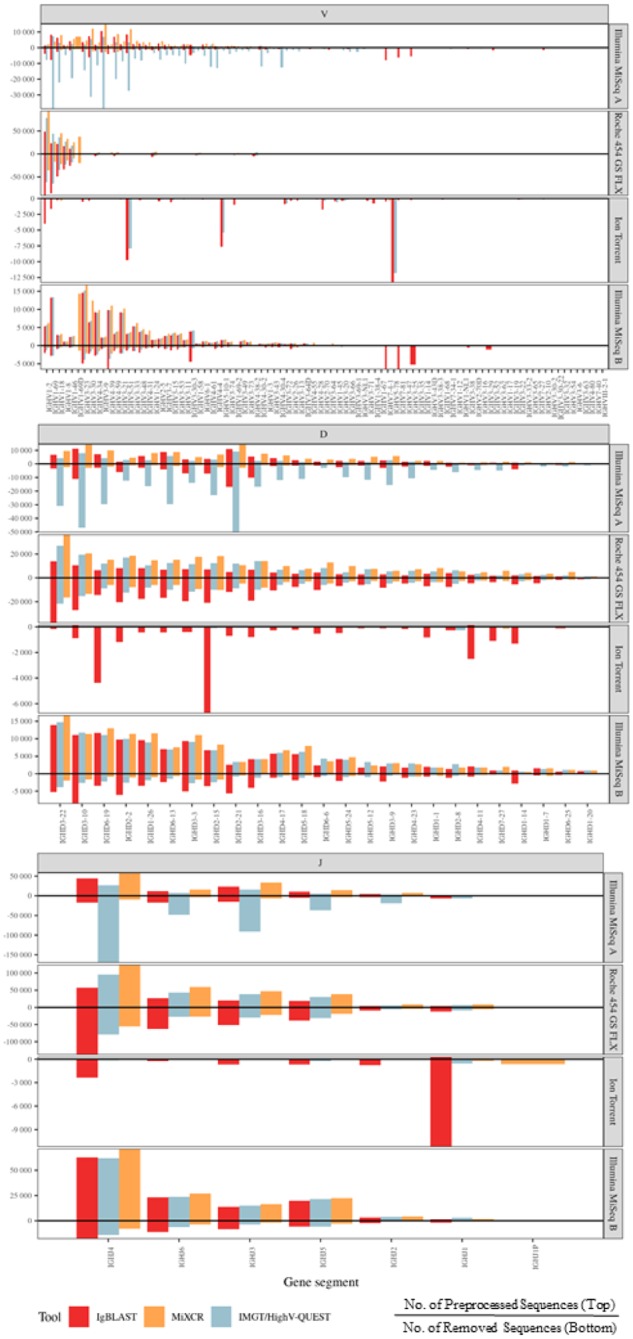
Alignment counts for each tool with experimental datasets. We compare the ability of each tool to align a given sequence to a particular gene (V, D and J). The hit counts before and after preprocessing and across different platforms (Illumina MiSeq, Roche 454 GS FLX and Ion Torrent) are shown. Preprocessing is defined as the removal of unproductive sequences (out-of-frame sequences, sequences containing premature stop codons, orphon genes or non-IgH sequences). Sequences that are removed by preprocessing by each tool are expressed by negative counts on the bottom while remaining preprocessed data is expressed on the top by positive counts. IgBLAST, red; MiXCR, orange; and IMGT/HighV-QUEST, blue. Only data for genes with total annotated count above the 30th percentile is shown
