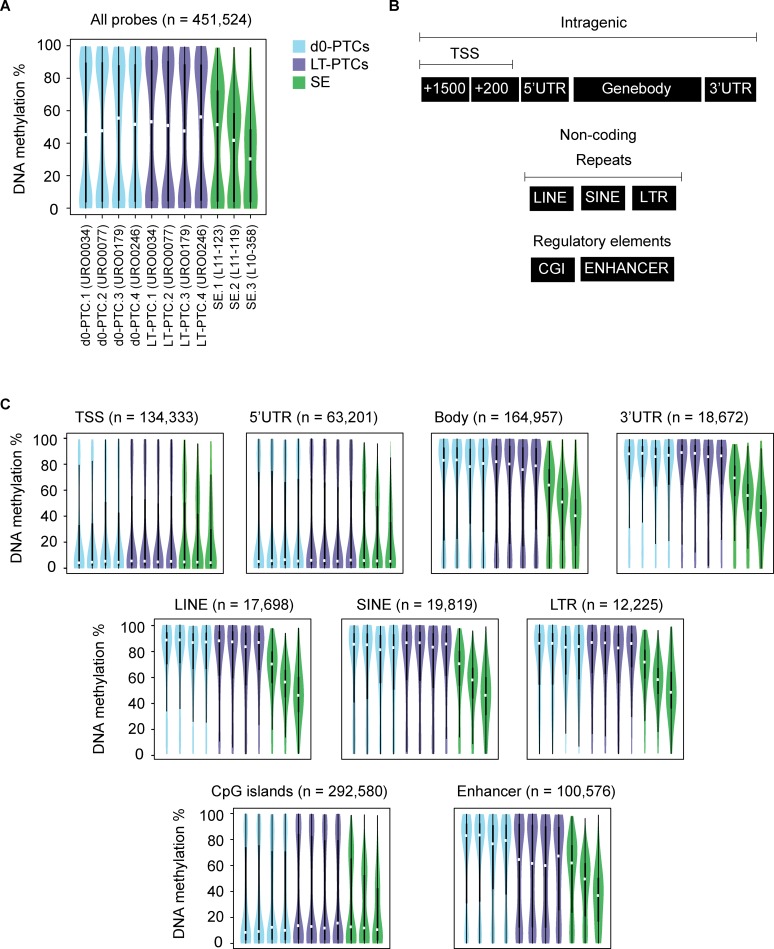
Fig 2. SE samples display a generally hypomethylated DNA methylation profile as compared to d0-PTCs and LT-PTCs.
(A) DNA methylation is deposited in a bipolar fashion in d0-PTCs and LT-PTCS, with most probes displaying a DNA methylation of either 0–10% or 90–100%, while a clear global hypomethylation is observed in SE. (B) Probe DNA methylation data was divided into nine genomic classes according to the genomic annotation of probes to the hg38 human genome reference build. A distinction was made between intragenic sequences (+1500bp of transcription start site (TSS), +200bp of TSS, 5' untranslated region (UTR), first exon, rest of genebody and 3'UTR) and non-coding sequences (long interspersed elements (LINE), short interspersed elements (SINE), long terminal repeats (LTR), CpG islands (CGI) and enhancer regions). (C) Distribution of DNA methylation over the nine genomic classes in d0-PTCs, LT-PTCS and SE.