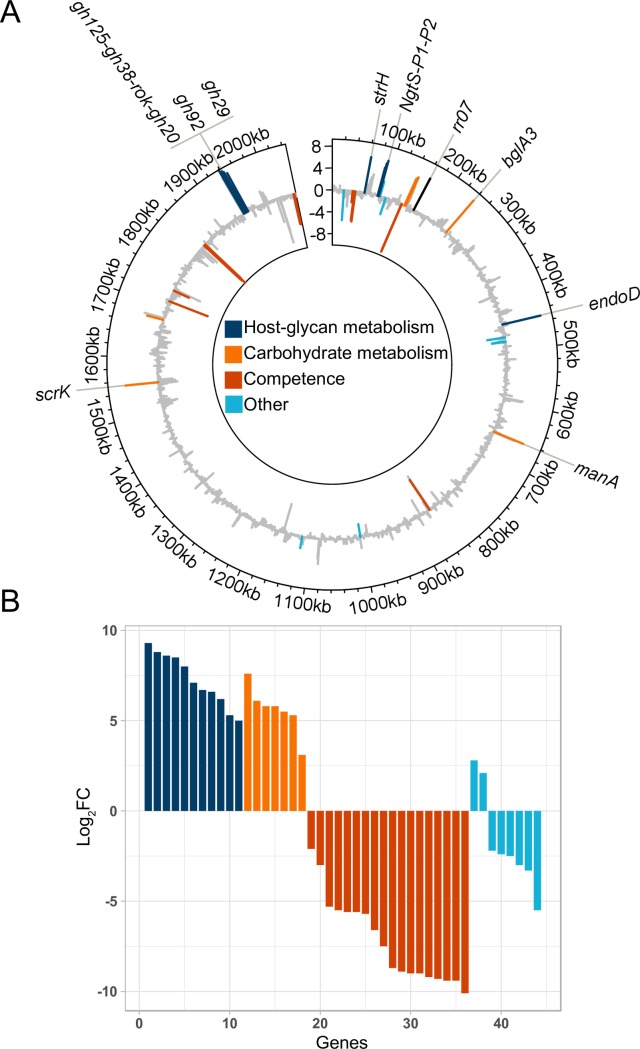
Fig 1. Overexpression of RR07 alters transcription of genes involved in host-glycan and carbohydrate metabolism.
The transcriptome of wild-type D39 and the isogenic RR07 overexpressing mutant was compared by RNA-seq. Strains were grown in quadruplicates in C+Y to OD600 = 0.5 at which point cells were harvested and RNA extracted. The RNA was prepared for and sequenced as described in materials and methods. Sequence reads were mapped to the genome of D39, normalized and quantified. Log2 fold changes (Log2FC), false-discovery rates (FDR) and reads per million (RPM) were calculated for each gene. Genes were considered upregulated if Log2FC > 2, FDR < 0.05 and RPM > 500 in the RR07 overexpressing strain. Genes were considered downregulated if Log2FC < -2, FDR < 0.05 and RPM > 50 in the wild type strain. A) Circular plot of D39 genome with log2FC at y-axis and genomic location at x-axis. Up- or downregulated genes are color coded according to known or predicted functions (grey: genes not considered regulated based on above criteria). B) Log2FC of up- or downregulated genes sorted according to predicted or known function. 11 genes involved in host-glycan metabolism and 7 in carbohydrate metabolism are all upregulated.