Abstract
Polybromo-1 (PBRM1) gene is a promising biomarker for immunotherapy in clear cell renal cell carcinoma. But to our knowledge, the frequency and clinical relevance of PBRM1 mutation in lung cancer remain unknown. We conducted a retrospective study to evaluate the prevalence of PBRM1 mutation and its correlation with preliminary response to immunotherapy in non-small cell lung cancer (NSCLC). Our results indicated that PBRM1 mutation was more likely to be a negative predictive biomarker for immunotherapy in NSCLC.
Subject terms: Predictive markers, Non-small-cell lung cancer, Cancer genomics
Introduction
Immune checkpoint blockade (ICB) therapy has been a pivotal treatment for lung cancer1,2. However, the response rate of cancer immunotherapy among lung cancer patients is still limited. Although several predictive biomarkers have been identified, such as PD-L1 expression, tumor mutation burden (TMB), and microsatellite instability, additional biomarkers should be found out to cover more patients who may benefit most from ICB therapy3–5.
Polybromo-1 (PBRM1), located on chromosome 3p21, is a tumor suppressor gene in many cancer types6. The existing knowledge regarding its function includes the control of cell cycle, promotion of genomic stability, apoptosis, centromeric cohesion, and so on7. Somatic mutations of PBRM1 are especially prevalent in clear cell renal cell carcinoma (ccRCC)8. Previous studies have found that PBRM1 mutation was a promising biomarker for immunotherapy in ccRCC9,10. Strong enrichment of immunostimulatory genes (including genes involved in hypoxia response and JAK–STAT signaling) may be the potential mechanism to enhance the response to ICB therapy in PBRM1-deficient ccRCC9. Compared with patients without the loss of PBRM1, patients with PBRM1 loss had longer overall survival (OS) and progression-free survival (PFS) (log-rank test P = 0.0074 and 0.029, respectively)9. To our knowledge, the frequency and clinical relevance of PBRM1 mutation in lung cancer remain unknown. Therefore, we conducted a retrospective study to evaluate the prevalence of PBRM1 mutation and its correlation with preliminary response to ICB therapy in non-small cell lung cancer (NSCLC).
Results
Patient characteristics
In the 2767 patients included in our study (Supplementary Fig. 1), PBRM1 mutation was identified in 84 NSCLC patients (3.04%, Supplementary Table 1). Fifty-one patients were found to have PBRM1 loss-of-function (LOF) mutation, accounting for 60.17% of the mutated patients (Supplementary Fig. 2). Among the 84 mutated patients, 56 (66.67%) had lung adenocarcinoma, and 23 (27.38%) had lung squamous cell carcinoma. The ratio of gender was balanced in this cohort (Male, 43, 51.19%; Female, 41, 48.81%). No significant difference in smoking status was observed between patients with PBRM1 mutation type (MT) and PBRM1 wild type (WT).
PBRM1 mutation predicts worse response to immunotherapy in NSCLC
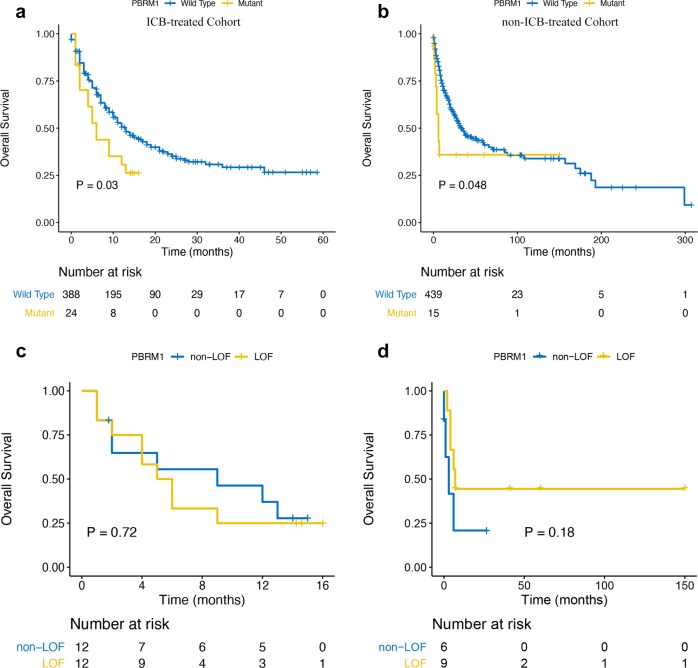
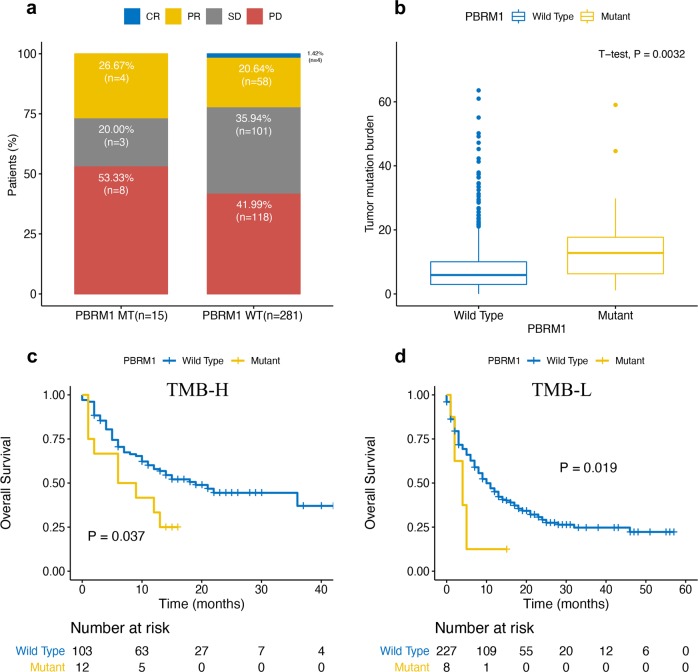
A combined cohort of 441 ICB-treated patients (385 from Memorial Sloan Kettering Cancer Center (MSKCC), 56 from Dana Farber Cancer Institute (DFCI)) were further analyzed to access the association between PBRM1 mutation and response to ICB therapy. As shown in Table 1, there was no significant difference in the distribution of gender, age, smoking status, and pathology between the two groups (P > 0.05). Most of the patients received anti-PD1/PD-L1 monotherapy, and the mean lines of treatment was about 2.3. In the cohort of ICB-treated patients (Nos = 412, PBRM1 MT = 24), the OS of the PBRM1-mutant patients was worse than that of those without the mutation (P = 0.03; Fig. 1a). The median OS of the 24 PBRM1-mutant patients was 6 months from the start of ICB therapy, while the median OS of the ICB-treated patients with PBRM1 WT was 13 months. To further investigate the role of PBRM1 mutation, we performed the multivariate Cox regression analysis including covariates (mono vs. combo therapy, lines of treatment, smoking, sex, age) using a 211 patients’ subgroup with available data. We found that the PBRM1 mutation was still negatively associated with poor OS (hazard ratio 2.16, 95% confidence interval 1.03–4.51, P = 0.041) after adjusting these covariates. A subgroup of 296 patients from ICB-treated cohort (N = 441) was with available data for the evaluation of response to ICB therapy (PFS, objective response rate (ORR), disease control rate (DCR) and durable clinical benefit (DCB)). Among them, 15 patients were detected with PBRM1 mutation. The median PFS was 2.1 months. The ORR was 26.67%, the DCR was 46.67%, and the DCB was 13.33% (Fig. 2a). In the cohort of non-ICB-treated patients (Nos = 454, PBRM1 MT = 15), there seems to be marginally significant difference in OS between the PBRM1 mutation subgroup and the PBRM1 WT subgroup, with the survival curves overlapped visually (P = 0.048; Fig. 1b).
Table 1.
The baseline characteristics of 441 ICB-treated patients.
| Characteristics | PBRM1 wild type (N = 415) | PBRM1 mutant (N = 26) | P value |
|---|---|---|---|
| Sex (%) | 0.207 | ||
| Male | 194 (46.7) | 16 (61.5) | |
| Female | 221 (53.3) | 10 (38.5) | |
| Age (%) | 0.199 | ||
| <31 | 1 (0.3) | 0 (0) | |
| 31–50 | 36 (9.9) | 1 (4.3) | |
| 50–60 | 80 (22.1) | 2 (8.7) | |
| 61–70 | 124 (34.3) | 7 (30.4) | |
| >71 | 121 (33.4) | 13 (56.5) | |
| NA | 53 | 3 | |
| Smoke (%) | 0.133 | ||
| Never | 60 (21.4) | 0 (0) | |
| Ever | 208 (74.0) | 14 (93.3) | |
| Current | 13 (4.6) | 1 (6.7) | |
| NA | 134 | 11 | |
| Pathology (%) | 0.697 | ||
| LUAD | 323 (77.8) | 21 (80.8) | |
| LUSC | 54 (13.0) | 2 (7.7) | |
| Other | 38 (9.2) | 3 (11.5) | |
| Therapy (%) | 0.210 | ||
| Mono | 377 (90.8) | 26 (100) | |
| Combo | 38 (9.2) | 0 (0) | |
| Lines of treatment (mean (SD)) | 2.24 (1.15) | 2.33 (0.89) | 0.775 |
Fig. 1. Kaplan–Meier curve comparing overall survival of patients whose tumors did or did not harbor PBRM1 mutations.
a The OS of the PBRM1-mutant patients was worse than that of those without the mutation in the cohort of ICB-treated patients. b There is marginally significant difference in OS between the PBRM1 mutation subgroup and the PBRM1 wild type subgroup in the cohort of non-ICB-treated patients. No survival difference between PBRM1 mutation types (LOF mutation and non-LOF mutation) were observed in the cohort of ICB-treated (c) and non-ICB-treated (d) patients. OS overall survival, ICB immune checkpoint blockade, LOF loss of function.
Fig. 2. PBRM1 mutation and response to ICB therapy.
a PBRM1 mutation and response to ICB therapy by 296 patients. b The boxplot showed that TMB was significantly higher in PBRM1-mutated patients. The boundary of the box closest to zero indicates the 25th percentile, a line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. The whiskers left and right of the box indicate the 90th and 10th percentiles. Points above and below the whiskers indicate outliers outside the 10th and 90th percentiles. Kaplan–Meier curve comparing overall survival of patients whose tumors did or did not harbor PBRM1 mutations in the TMB-High (c) and TMB-Low (d) subgroups. ICB immune checkpoint blockade, TMB, tumor mutation burden.
In addition, we also observed that there was no survival difference regarding MTs (LOF mutation and non-LOF mutation), when considering the OS in the ICB and non-ICB-treated cohort (Fig. 1c, d).
TMB was significantly higher in PBRM1-mutated patients compared with that in PBRM1 WT patients (median 12.79 and 5.9, respectively, P < 0.05, Fig. 2b). In the TMB-high and TMB-low subgroup analysis, we still observed the worse OS in the PBRM1-mutant patients treated with ICB therapy (P < 0.05, Fig. 2c, d).
Discussion
In this retrospective study, we combined data from three institutions to investigate the clinical efficacy of ICB therapy in NSCLC patients with or without PBRM1 mutation. Unlike ccRCC, NSCLC seemed to follow a different PBRM1 mutation pattern. The prevalence of PBRM1 mutation (NSCLC: 84/2767, 3.04%; ccRCC: 162/402, 40.30% in The Cancer Genome Atlas (TCGA)) and the proportion of truncating mutation (NSCLC: 51/84, 60.17%; ccRCC: 144/162, 93.51% in TCGA) were relatively low in NSCLC (Supplementary Fig. 2). Our findings suggested that PBRM1-mutant NSCLC patients might get less survival benefit from ICB therapy, unlike previously reported data in ccRCC. Interestingly, PBRM1-mutant patients tended to have higher TMB. But no matter in TMB-high subgroup or in TMB-low subgroup, PBRM1-mutant patients who received ICB therapy had worse survival than those without PBRM1 mutation. Besides, PBRM1 mutation was not a remarkable prognostic factor in NSCLC patients according to our analysis in non-ICB-treated patients. Taken together, our results indicated that PBRM1 was more likely to be a negative predictive biomarker for ICB therapy in NSCLC.
To our knowledge, our study was the first study to estimate the role of PBRM1 mutation in both ICB and non-ICB-treated NSCLC cohorts. However, due to data restrictions, not all patients have the full record of clinical data. There was discrepancy in the patients when performing different analysis. We were also not able to include PD-L1 level, microsatellite instability and other factors that might influence the response to ICB therapy in our analysis. In addition, the number of PBRM1-mutated patients was limited, this low frequency may limit the utility of PBRM1 mutation as a predictive biomarker, and we still have to interpret the results with caution. Moreover, PBRM1 mutation did not help predict benefit from the first-line ICB treatment for ccRCC. Most patients received ICB therapy as second or later-line therapy in our cohort. It is still unknown whether PBRM1 mutation can be a predictive biomarker for the first-line ICB therapy. Therefore, further prospective research is warranted to confirm the negative predictive role of PBRM1 in NSCLC ICB therapy.
Methods
Patients
We analyzed the combined NSCLC cohort of 2767 patients, from three sources: (1) TCGA (N = 1144), (2) MSKCC (N = 1567), and (3) DFCI (N = 56)5,11–14.
PBRM1 mutation
We first estimated the prevalence of PBRM1 mutation in the whole NSCLC cohort. PBRM1 mutation was defined as any SNV or indel, including putative truncating mutations (nonsense mutations, frameshift, insertions and deletions, and splice-site mutations) and other alterations presumed not to be truncating (In-frame insertions and deletions, missense mutations etc.). Notably, homozygous deletion was also calculated in the PBRM1 mutation. Moreover, we classified PBRM1 mutations into two type: LOF (any truncating mutation and homozygous deletion) and non-LOF.
PBRM1 mutation and response to immunotherapy
A subset of ICB-treated patients (N = 441, 385 from MSKCC, 56 from DFCI) with annotated clinical records were further analyzed for the association between PBRM1 mutation and response to ICB therapy. The OS, PFS (calculated from the date of first ICB infusion) and response to ICB therapy (ORR, DCR, and DCB) were evaluated among these 441 ICB-treated patients. We also calculated the OS (calculated from the date of first chemotherapy infusion) of 454 non-ICB-treated patients from MSKCC cohort. The results of subgroup analysis were also displayed according to the status of PBRM1 LOF mutations. In addition, we investigated the relationship between TMB and PBRM1 mutation status (804 available patients, 454 non-ICB; 350 ICB). In order to further clarify the role of PBRM1 mutation, we classified the ICB-treated patients into two groups (TMB-High and TMB-low, cut-off data: TMB = 10 mut/Mb), and compared their OS. We also showed the PBRM1 mutation landscape of 402 patients with ccRCC from the TCGA database. Institutional review board approval and informed consent were waived because all data were de-identified and publicly available.
Statistical analysis
Patients’ characteristics at baseline and response to therapy were compared by T test or Mann–Whitney U test (continuous variables) and Pearson chi‐squared test (categorical variables). Kaplan–Meier curve was used to describe the OS and PFS, and the differences between groups were tested by log‐rank method. All statistical analyses were performed using R version 3.5.3 software (Institute for Statistics and Mathematics, Vienna, Austria; www.r-project.org). Statistical significance was set at two-sided P < 0.05.
Supplementary information
Acknowledgements
This work was supported by: National Key R&D Program of China (Grant nos. 2016YFC0905500, 2016YFC0905503), Chinese National Natural Science Foundation project (Grant nos. 81872499, 81772476), Science and Technology Program of Guangdong (Grant no. 2017B020227001), and Science and Technology Program of Guangzhou (Grant nos. 201607020031, 201704020072). The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The authors acknowledge the efforts of The Cancer Genome Atlas (TCGA), Memorial Sloan Kettering Cancer Center (MSKCC), and Dana Farber Cancer Institute (DFCI) in providing high-quality data resources for researchers.
Author contributions
H.Z., J.L., Y.Z., Y.H., and J.S. contributed equally to this work and should be considered as co-first authors. Study concept and design: W.F., H.Z., Y.Z., Y.H., and L.Z. Acquisition, analysis, or interpretation of data: All authors. Drafting of the manuscript: H.Z., J.L., and L.Z. Critical revision of the manuscript for important intellectual content: W.F., Y.Z., Y.Y., Y.H., and L.Z. Statistical analysis: H.Z., J.L., and Y.Z. Obtained funding: W.F., Y.H., L.Z. Study supervision: L.Z.
Data availability
The data that support the findings of this study are available from the website [cBioPortal for Cancer Genomics] (https://www.cbioportal.org/), and are also available from the corresponding author on reasonable request. TCGA: Pan-Lung Cancer11 [https://www.cbioportal.org/study/summary?id=nsclc_tcga_broad_2016] MSKCC: MSK-IMPACT Clinical Sequencing Cohort12 [https://www.cbioportal.org/study/summary?id=msk_impact_2017] DFCI: MSS Mixed Solid Tumors13 [https://www.cbioportal.org/study/summary?id=mixed_allen_2018].
Code availability
The code that support the findings of this study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Huaqiang Zhou, Jiaqing Liu, Yaxiong Zhang, Yan Huang, Jiayi Shen.
Contributor Information
Wenfeng Fang, Email: fangwf@sysucc.org.cn.
Li Zhang, Email: zhangli6@mail.sysu.edu.cn.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41698-020-0112-3.
References
- 1.Gandhi L, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 2018;378:2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 2.Paz-Ares L, et al. Pembrolizumab plus chemotherapy for squamous non–small-cell lung cancer. N. Engl. J. Med. 2018;379:2040–2051. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 3.Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer. 2019;7:278. doi: 10.1186/s40425-019-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PloS ONE. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mota ST, et al. New insights into the role of polybromo-1 in prostate cancer. Int. J. Mol. Sci. 2019;20:2852. doi: 10.3390/ijms20122852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miao D, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359:801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun DA, et al. Clinical validation of PBRM1 alterations as a marker of immune checkpoint inhibitor response in renal cell carcinoma. JAMA Oncol. 2019;5:1631–1633. doi: 10.1001/jamaoncol.2019.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell JD, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao D, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018;50:1271–1281. doi: 10.1038/s41588-018-0200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizvi H, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J. Clin. Oncol. 2018;36:633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the website [cBioPortal for Cancer Genomics] (https://www.cbioportal.org/), and are also available from the corresponding author on reasonable request. TCGA: Pan-Lung Cancer11 [https://www.cbioportal.org/study/summary?id=nsclc_tcga_broad_2016] MSKCC: MSK-IMPACT Clinical Sequencing Cohort12 [https://www.cbioportal.org/study/summary?id=msk_impact_2017] DFCI: MSS Mixed Solid Tumors13 [https://www.cbioportal.org/study/summary?id=mixed_allen_2018].
The code that support the findings of this study are available from the corresponding author on reasonable request.