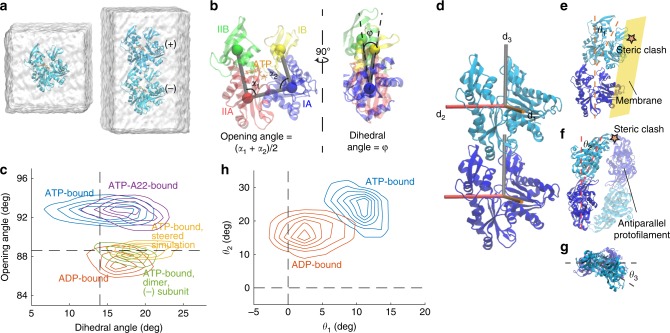
Fig. 2. MreB monomer and dimer conformations are nucleotide dependent.
a Simulated systems of an MreB monomer (left) and a single protofilament with two subunits (“2 × 1 protofilament”, right). Each MreB subunit is bound to a nucleotide, with the whole system surrounded by a water box. In the 2 × 1 single protofilament, we refer to the top and bottom MreB subunits as the (+) and (−) subunits, respectively. b Definitions of opening angle and dihedral angle for an MreB monomer, with the centers-of-mass of the four subdomains shown as colored spheres. c Contour density plot of the distributions of opening and dihedral angles for each simulation system from the last 40 ns of the simulation. MreB subunits essentially adopted one of two conformations in simulations. ATP-bound MreB monomers exhibited large opening angles in the presence (purple) and absence (blue) of A22, while an ADP-bound monomer (red) and the (−) subunit of an ATP-bound dimer (green) had smaller opening angles. Steering of the opening angle of an ATP-bound monomer to its value in the crystal structure (yellow) mimicked the conformation of an ADP-bound monomer. Dashed lines denote the values of the opening and dihedral angles in the crystal structure (PDB ID: 4CZF). d An MreB dimer from a single protofilament, with three axes (d1, d2 and d3) overlaid on each subunit that were used to compute the degree of bending and twisting between them. e) Illustration of θ1, with positive θ1 denoting bending toward the membrane surface (yellow). Non-zero θ1 leads to a steric clash with the membrane surface. f Illustration of θ2, with positive θ2 denoting bending toward the inter-protofilament interface. The paired antiparallel protofilament is shown in semi-transparency. Non-zero θ2 leads to a steric clash with the paired protofilament. g Illustration of θ3 from the top of a protofilament, with positive θ3 denoting left-handed twisting. h Contour density plot for the distributions of θ1 and θ2 from the last 40 ns (200 frames) of the simulations, with both ATP- and ADP-bound single protofilaments bending toward the membrane side and toward the inter-protofilament interface. An ATP-bound single protofilament exhibited more substantial bending in both directions than an ADP-bound single protofilament. Dashed lines denote the respective angles in the crystal structure.