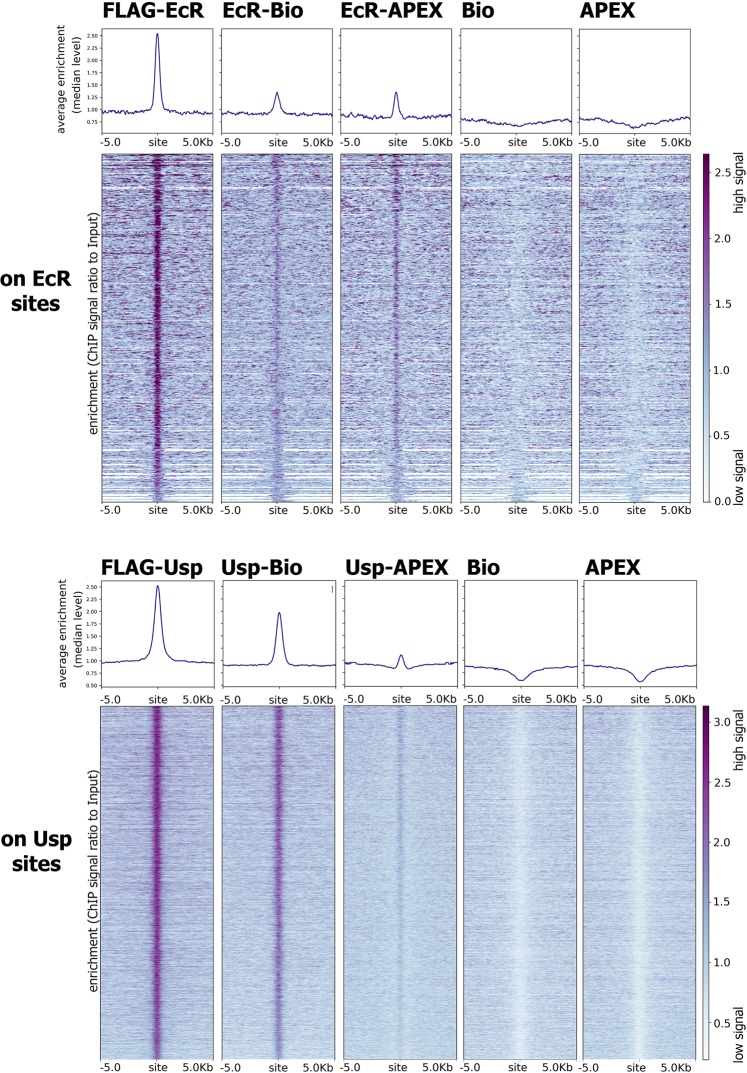
Figure 1.
EcR and Usp fused to APEX2 and BioID2 enzymes bind genomic sites of EcR and Usp. (A) Distribution of EcR-BioID2 (EcR-Bio), EcR-APEX2 (EcR-APEX), BioID2 (Bio) and APEX2 (APEX) on EcR-bound sites (which were defined using control Drosophila S2 cell line expressing EcR in relation to the corresponding input DNA) in Drosophila S2 cells after 1-hour treatment with 20E. (B) Distribution of Usp-BioID2 (Usp-Bio), Usp-APEX2 (Usp-APEX), BioID2 (Bio) and APEX2 (APEX) on Usp-bound sites (which were defined using control Drosophila S2 cell line expressing Usp in relation to the corresponding input DNA) in Drosophila S2 cells after 1-hour treatment with 20E. Protein binding level was estimated by ChIP-Seq using anti-FLAG antibodies. Heatmaps represent enrichment of ChIP-Seq signal over Input DNA (presented as the ratio of the number of reads at a given point). Pile-up profiles were calculated as a median of binding levels of corresponding proteins in a given distance out from the binding site.