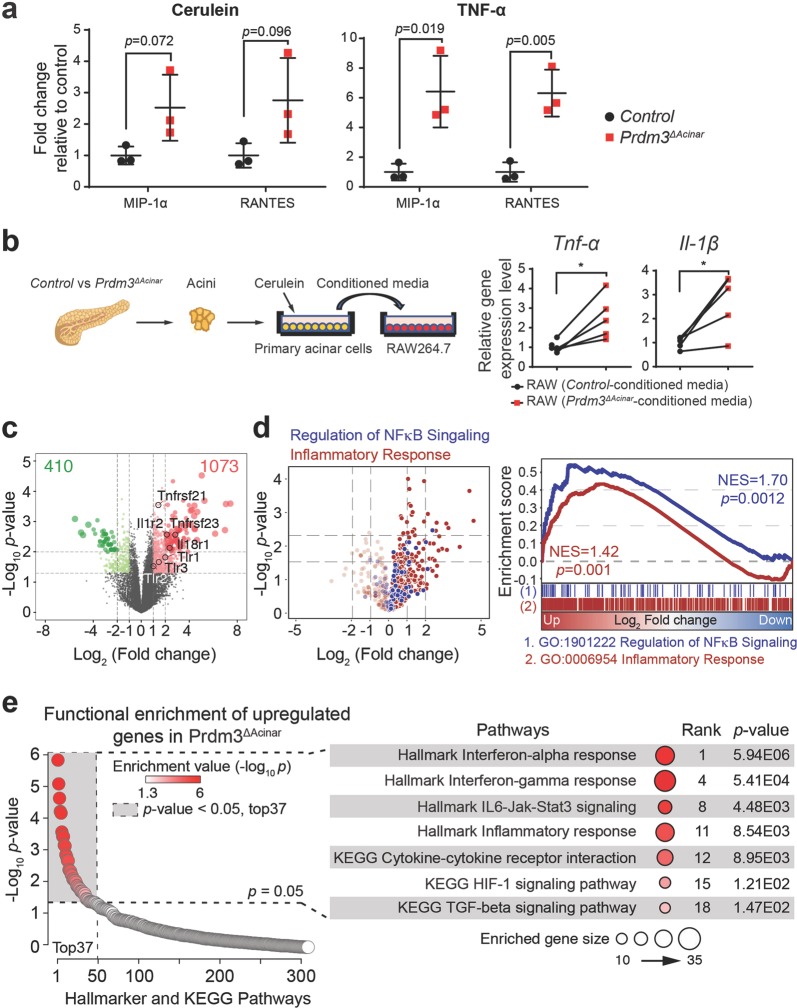
Fig. 5. Inactivation of Prdm3 enhances inflammatory response pathways.
a Cytokine secretion was determined using a Raybiotech mouse inflammation array. Primary acinar cells, isolated from control (n = 3) and Prdm3ΔAcinar (n = 3) mice, were exposed to cerulein (10 nM) or TNF-α (100 ng/ml) for 6 h. Levels of cytokines (MIP-1α and RANTES) in culture medium were expressed relative to control. p-values were calculated based on two-tailed t-test. b Primary acinar cells were isolated from Prf1aCreER (n = 5) vs. Prdm3ΔAcinar (n = 5) mice and cultured in Waymouth complete media in the presence of 10 nM cerulein for 6 h. Raw264.7 cells were treated with the above acinar-cell-conditioned media for 16 h and collected for quantitative real-time PCR for the indicated cytokines. Each set of connected dots in b depicts an independent biological replicate (n = 5 experiments). Data show mean ± SD. Statistical analysis: Two-tailed t-test. *p < 0.05. c Volcano plot showing a total of 1483 differentially expressed genes in primary acinar cells isolated from Prdm3ΔAcinar mice, relative to control Ptf1aCreER. Individual genes are labeled and circled in black. d Volcano plot showing differentially expressed genes belonging to inflammatory responses and NF-κB signaling are highlighted in red and blue, respectively. Gene Set Enrichment Analysis (GSEA) of differentially expressed genes (Ptf1aCreER vs. Prdm3ΔAcinar) identified enrichment of immune responses and regulation of NF-κB signaling. Normalized enrichment score (NES) and p-values are shown. e Functional annotation on 1073 upregulated genes in primary acinar cells isolated from Prdm3ΔAcinar mice, relative to control. Significant KEGG and Hallmark terms, p-values and ranks are shown.