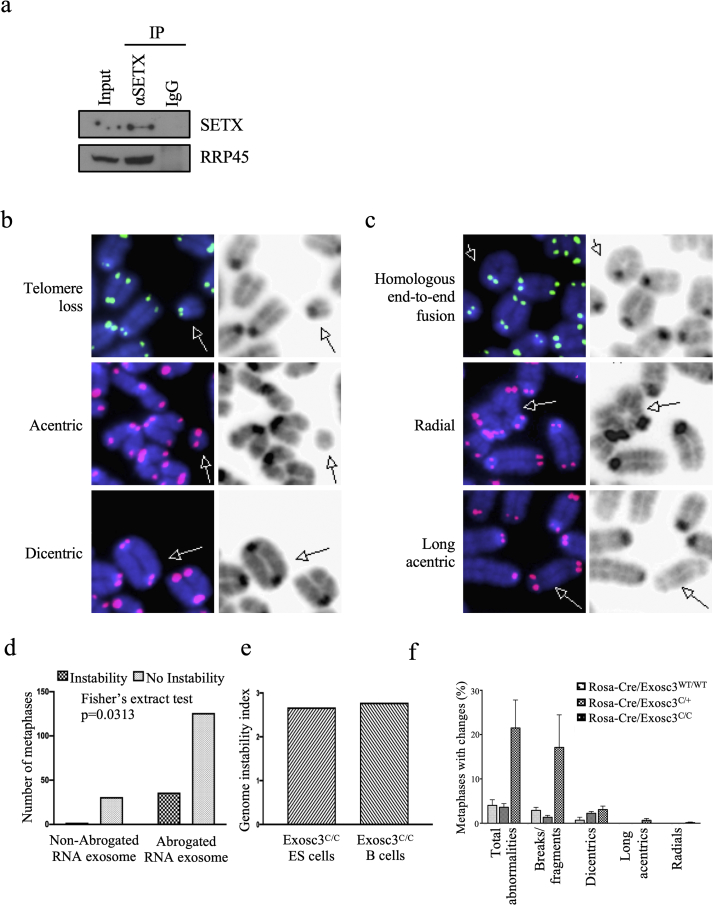
Figure 1.
Senataxin-interacting RNA exosome complex is important for maintenance of B-cell genomic integrity. (a) Human Burkitt's lymphoma Ramos cells were harvested, lysed, and prepared for SETX immunoprecipitation. Input (1/15 of lysate used for IP and normal mouse IgG1 controls are displayed, along with Western blotting using antibodies against RNA exosome core subunit RRP45. Full images are in the supplemental figure 1a. (b) Structural abnormalities in RNA exosome-deficient stimulated B cells. Examples are shown of telomeric telomere loss, acentric chromosomal fragments, and dicentric chromosomes. These abnormal chromosomal structures accrue at a higher degree in activated ROSA26Cre-ERT2/+; EXOSC3COIN/COIN B lymphocytes than in their RNA exosome-competent counterparts. Structures are shown with Cy3 (green) or Cy5 (red) fluorophore-tagged peptide nucleic acid (PNA) telomeric probes and 4′,6-diamidino-2-phenylindole (DAPI, blue) staining (on the left) and with transmitted light (on the right). Densities are indicative of centromeres. Chromosomal fragments without centromeres are acentric (middle row), while chromosomal structures with 2 centromeres are dicentric (bottom row). Fluorophore-labeled telomeric probes denote the presence or absence of telomeres (centromeric or telomeric telomeres; seen here is telomeric telomere loss in the top row). (c) Additional representations are shown for homologous end-to-end fusion, radial, and long acentric chromosomes. These abnormal chromosomal structures accrue at a higher degree in activated ROSA26Cre-ERT2/+; EXOSC3COIN/COIN B lymphocytes than in their RNA exosome-competent counterparts. Note the chromatid fusion and absence of telomeric telomeres in the top picture pair; the middle pair shows a quadriradial chromosome; the bottom pair shows a long acentric chromosome. Cognizant that in 2 mouse chromosomes lack of a centromere is expected, this chromosome is excessively long to be one of those. (d) Comparison of the percentage of fixed metaphases with chromosomal abnormalities before and after RNA exosome abrogation in embryonic stem (ES) cells. There is a statistically significant increase in the number of metaphases with genomic instability in the RNA exosome-abrogated background compared to the non-abrogated RNA exosome background (Fisher's exact test, p = 0.0313). (e) Genomic instability indices devised to compare the level of genomic instability observed when RNA exosome activity is impaired in both ESCs and B cells. Relative to their exosome-sufficient baselines, both ES and B cells have increased genomic instability upon abrogation of RNA exosome function. The genomic instability index [(D–S)/S] normalizes the change in instability seen in RNA exosome-deficient backgrounds (“D-S”) to baseline instability levels observed in RNA exosome-sufficient backgrounds (“S”) in our assay. (f) Breakdown of chromosomal instability events in activated B cells. A compilation of the different phenotypes is plotted for the three backgrounds. For all abnormalities, we observed a statistically significant difference between the RNA exosome-deficient B cells and the RNA exosome-competent backgrounds: ANOVA, p = 0.0409, R2 = 0.5989. 4 animals were used for the WT background, 2 for the heterozygous background, and 4 for the ROSA26Cre-ERT2/+; EXOSC3COIN/COIN background. The respective number of metaphases analyzed is indicated for each mouse background.