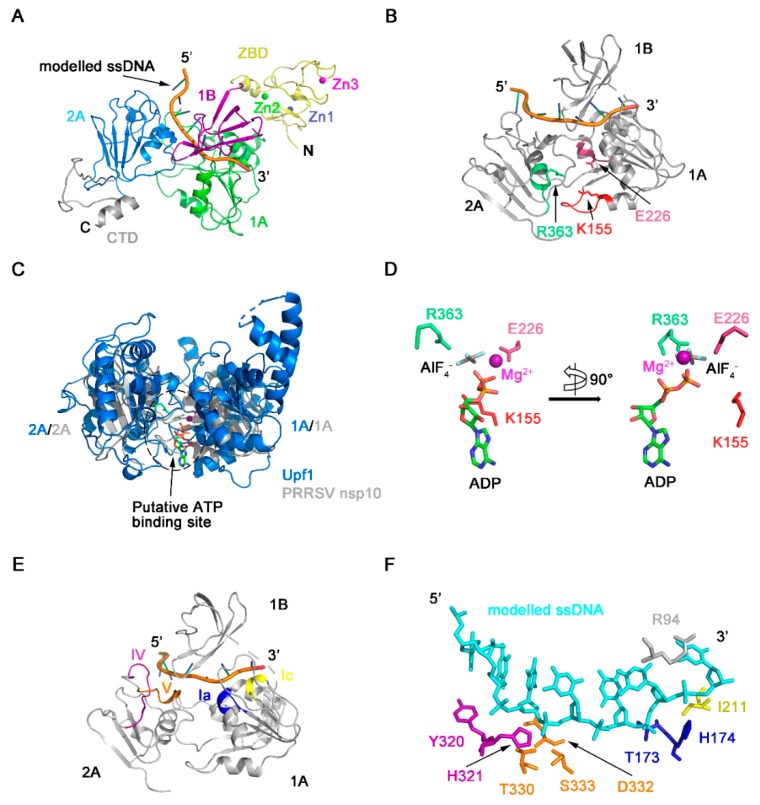
Figure 6.
Model of PRRSV nsp10-DNA complex. (A) Model of PRRSV nsp10-DNA complex was generated based on the structure of EAV nsp10-DNA complex. The rest part of ssDNA is omitted for clarity. Domain colors are the same as in Figure 1A. (B) Position of conserved motifs and key residues necessary for NTP binding and hydrolysis in the structure of the nsp10-DNA complex model. (C) Superposition between helicase cores of human Upf1-ADP-AlF4- (PDB ID: 2XZO) in marine and PRRSV nsp10-DNA complex model in grey. These two structures are superimposed on 2A domains. (D) Close-up view of the modelled ADP-AlF4- with the residues of PRRSV nsp10, which involved in NTP binding and hydrolysis. A 90° rotation view is shown in the right panel. Motifs colors are the same as in Figure 5A. (E) Position of conserved motifs necessary for nucleotide binding in the structure of the nsp10-DNA complex model. The ZBD, the CTD, and the rest part of ssDNA in the complex model are omitted for clarity. (F) Close-up view of the modelled nucleic acids with the residues involved in nucleic acids’ binding. Motifs colors are the same as in Figure 5A.