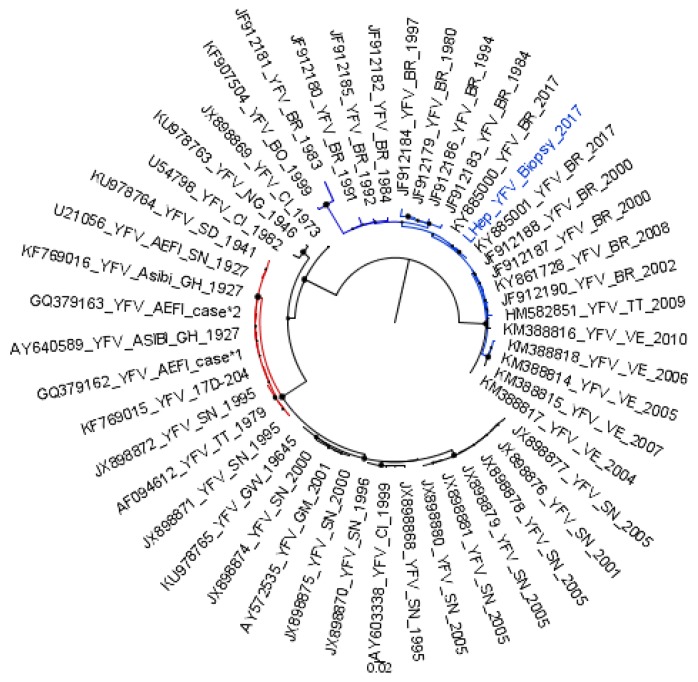
Figure 2.
Detection and phylogeny analyzis of YFV RNA sequences in the liver biopsy collected at the 93rd day post-onset of first symptoms of yellow fever. A sub-tree from the maximum-likelihood tree inferred using Yellow fever virus sequences (211 nucleotides) is shown. YFV strains from South American genotypes are shown in blue. The YFV detected in the liver biopsy (highlighted in blue) is grouped within the South American clade, apart from the vaccine strains and vaccine-related strains (red). The YFV strains belonging to African genotypes are shown in black, except vaccine and vaccine-related strains, which are colored in red. YFV strains are identified by their Genbank identification number, and by the year they were detected. The bootstrap values are represented by circles drawn in scale in the nodes. The scale indicates nucleotide substitution per site. The tree was reconstructed using the Kimura-2 parameters nucleotide substitution model with gamma distribution (four categories), and analyzes were performed using MEGA7 [12].