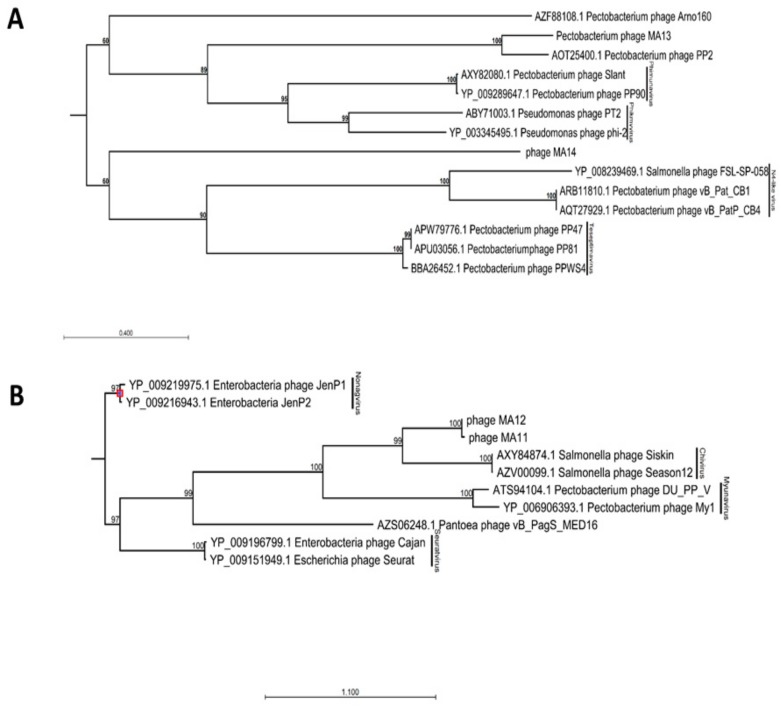
Figure 2.
Maximum-likelihood phylogenetic analysis of major capsid proteins of phages isolated in this study constructed using a neighbor–joining method with protein substitution model Dayhoff (PAM) using CLC Genomic Workbench 9.5.4. (A) Members of Podoviridae family phages. (B) Members of Siphoviridae family phages. For phages obtained from GenBank capsid proteins accession numbers followed by phages names; phages isolated in this study: phage MA11, phage MA12, phage MA13 and phage MA14. Bootstrap probabilities values < 50% were collapsed.