Abstract
The development, progression, recurrence, and metastasis of hepatocellular carcinoma (HCC) are closely associated with an abnormal liver-regenerating microenvironment (LRM). Therefore, preventing and reversing an abnormal LRM is a potential therapeutic strategy against HCC. Studies are increasingly focusing on the impact of regeneration, fibrosis, angiogenesis, inflammation, immunomodulation, and hepatic stem cells on HCC development and progression. As a key epigenetic mechanism, DNA methylation is extensively involved in regulating physiological and pathological pathways. In this review, we summarize recent findings on the role of DNA methylation in the fibrotic, angiogenic, inflammatory/immune, and stem cell microenvironments of HCC, and discuss new advances in Traditional Chinese Medicine (TCM) on influencing the abnormal LRM, so as to gain new insights into alleviating the abnormal LRM via regulating DNA methylation by TCM.
MeSH Keywords: Carcinoma, Hepatocellular; Cellular Microenvironment; DNA Methylation; Liver Regeneration; Medicine, Chinese Traditional
Background
Hepatocellular carcinoma (HCC) is an aggressive malignancy characterized by undetectable onset, high morbidity and mortality, and high annual increase rate [1–3]. In-depth understanding of HCC pathogenesis is key to improving prevention and treatment. Recent studies have revealed multiple facets of the relationship between HCC pathogenesis and the tumor microenvironment. In particular, the mechanisms underlying the development of an abnormal liver-regenerating microenvironment (LRM) offer new strategies of HCC prevention and treatment [4,5].
Basic Concept of DNA Methylation
Although the exact pathogenesis of HCC is still unclear, studies show an essential role of epigenetics in HCC development and progression [6]. Epigenetic modification involves changes in gene expression without altering the DNA sequence, and is stably inherited intra- or inter-generationally [7]. DNA methylation was the first epigenetic mechanism to be identified, and usually affects the cytosine residues of CpG dinucleotides (the CpG-rich regions in the genome are typically 300–3000 bp long). DNA methyltransferase adds a methyl group to the C5 of cytosine, and forms a stable 5-methylcytosine structure. Over 60% of the CpG regions in the mammalian genome are methylated, and the unmethylated CpG regions are known as CpG islands. Since the location of most CpG islands coincides with gene promoter regions and transcription initiation sites [8], their methylation is negatively associated with gene expression levels [9]. Therefore, temporal changes in DNA methylation are inextricably linked to the development, evolution, diseases, and death of living organisms.
LRM of HCC
HCC is a multifactorial, multi-step, multi-gene, and multi-mutation disease. In recent years, studies on HCC pathogenesis, prevention, and treatment have gradually shifted from a focus on the cancer cells to the HCC microenvironment [5,10,11]. Liver regeneration and repair is critical for post-disease recovery, but is severely impaired during specific pathological conditions wherein the LRM is adversely affected [12]. Since an abnormal LRM is a contributing factor for HCC development, progression, and metastasis [4,13], studies are increasingly focusing on the impact of regeneration, fibrosis [14], angiogenesis [15], inflammation, immunomodulation [16], and hepatic stem cells [17] on HCC development and progression. This in turn has led to the development of new LRM-based prevention and treatment strategies for HCC [18], as well as tertiary protocols targeting both HCC cells and LRM [19]. Various therapeutic modalities, including surgical resection, radiation therapy (RT), and chemotherapy, that can eliminate HCC cells and restore the LRM have shown encouraging results in clinical studies. The development of HCC cell- and LRM-targeting treatment regimens can significantly improve HCC prevention and treatment [20–22].
LRM of HCC and DNA Methylation
Abnormal LRM and DNA methylation patterns are mutually dependent; while an abnormal LRM can promote the methylation of CpG islands in key anti-tumor pathways/genes and downregulate their expression levels, DNA methylation can accelerate the formation and aggravation of an abnormal LRM, thereby facilitating HCC progression. Since fibrotic, angiogenic, inflammatory/immunological, and stem cell microenvironments of HCC can all play a role in the generation of an abnormal LRM, targeting the specific DNA methylation changes can be a potential anti-HCC strategy.
DNA methylation and liver fibrosis
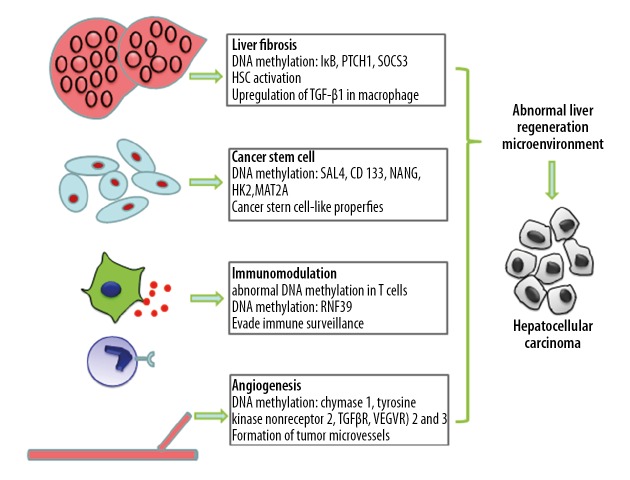
Hepatic fibrosis/cirrhosis, a common pathological change accompanying chronic liver diseases and HCC, is a result of an imbalance between extracellular matrix (ECM) synthesis and degradation. A persistent fibrotic microenvironment is conducive to the initiation and exacerbation of HCC, and is closely associated with HCC cell proliferation, metastasis, and drug resistance [23]. Hepatic stellate cell (HSC) activation is a central event in the development, progression, and exacerbation of hepatic fibrosis, which significantly depends on its methylome. DNA hypermethylation at specific sites was reported to downregulate IκB expression in activated HSCs to induce a fibroblast-like transition, which was reversed upon exposure to a demethylating agent [24]. HSC activation is also closely associated with PTCH1 hypermethylation, and both MeCP2 knockdown and methylation inhibitors increased PTCH1 expression and inhibited HSC activation [25]. Abnormal HSC methylation not only affects CD133, Notch1, and Notch3 expression, but also inhibits HCC stem cell differentiation [26]. Abnormal liver macrophage methylation has also been implicated in the formation of the fibrotic microenvironment [27]. Ogata et al. showed that increased methylation of SOCS3 downregulated its expression in tumor and tumor-free regions of the liver, and promoted STAT3-mediated upregulation of TGF-β1 and formation of a fibrotic microenvironment [28]. Taken together, evidence shows that abnormal DNA methylation in liver stromal cells promotes a fibrotic microenvironment, which leads a vicious cycle of abnormal DNA methylation and aggravated fibrosis.
DNA methylation and angiogenesis
Dysregulated liver regeneration-associated cytokines and their receptors in the abnormal LRM are known to affect angiogenesis in HCC, leading to the formation of a microcirculatory system that facilitates HCC cell proliferation and metastasis. When the diameter of a solid tumor exceeds 1 mm, an intra-tumoral microvascular network is formed to transport nutrients to the cancer cells [29]. Endothelial cells (ECs) are the building blocks of vascular structures, and the migration and maturation of EC precursors is controlled by genetic reprogramming. In addition, cancer stem cells (CSCs) can also differentiate into ECs for tumor microvascular formation [30]. The ECM not only protects the ECs, but it also secretes a large amount of pro-angiogenic factors [31]. Recent studies show that cancer cells can also aggregate into vascular-like structures to increase blood supply to the tumor tissues, a phenomenon known as vasculogenic mimicry (VM), even in the absence of ECs [32]. Changes in DNA methylation significantly affect neo-angiogenesis in HCC. Studies show that hypomethylation promotes tumor microvasculature formation and metabolism by upregulating chymase 1, tyrosine kinase nonreceptor 2, and transforming growth factor beta receptor II (TGFβR) in HCC cells [33,34]. The vascular endothelial growth factor receptors (VEGFR) 2 and 3 are also regulated by DNA methylation to enable the formation of tumor microvessels for tumor cell survival, proliferation, and metastasis [35].
DNA methylation and immunomodulation
Liver tissue inflammation induced by abnormal immune response is a trigger for liver regeneration and repair. Repeated inflammatory damage leads to continuous regeneration and repair, which increases the risk of abnormal liver regeneration. Furthermore, abnormal regeneration and repair increases the production of regenerative and inflammatory factors, resulting in a vicious cycle of inflammatory injury and LRM abnormality. HBV infection is the most common trigger of HCC, and is regulated by the immune response to the virus [36]. The liver immunological niche is not only linked to the development of HCC, but also affects its progression, metastasis, and post-treatment recurrence. A previous study showed that Tregs are more abundant in HCC compared to non-tumor tissues, and that the high percentage of peripheral Tregs in HCC patients is negatively correlated with HCC prognosis [37]. HBV-induced chronic hepatitis can lead to abnormal DNA methylation in hepatocytes [38,39], as well as in the peripheral monocytes and T cells of HCC patients [40]. NKG2D receptor plays an important role in protecting the host from infection and cancer. By recognizing the ligands induced on the infected or tumor cells, NKG2D regulates the activation of lymphocytes and enhances immunity, thus eliminating the cells expressing ligands [41]. Some studies have shown that the DNA methylation pattern is related to expression of the NKG2D gene, and the frequency of NKG2D promoter methylation in HCC patients is significantly higher than that in chronic hepatitis B patients and healthy people [42]. Later studies also found that in patients with liver cancer, due to the hypermethylation of NKG2D promoter, gene expression was silenced, mRNA expression level was reduced, and the expression of NKG2D on the cell surface was reduced, thereby weakening the killing function of tumor cells [43]. Abnormal methylation of RNF39, a structural component of the MHC complex that is essential for antigen presentation and processing, is associated with HCC immune evasion [38]. The ability of tumor cells to evade immune surveillance and maintain a high rate of proliferation is closely associated with immune suppression. Together, these studies show that abnormal DNA methylation influences development of the HCC immune microenvironment. However, the correlation between the altered methylome of specific immune cells and their functions remains to be further investigated.
DNA methylation and CSCs
The cancer stem cell hypothesis is now widely accepted by oncologists, and HCC stem cells are identified on the basis of surface CD44 and CD90 [44]. Studies show that the differentiation of mesenchymal stem cells (MSC) and hematopoietic stem cells (HSC) is associated with HCC development and progression [45,46]. In the normal LRM, stem cells can differentiate into hepatocytes and enable tissue regeneration, while the abnormal LRM triggers their transformation into HCC stem cells, which are the major source of cancer cells. The uncontrolled proliferation of these CSCs further exacerbates the abnormal LRM. The transformation of normal hepatic stem cells to CSCs is epigenetically regulated. Raggi et al. found that DNA methylation-mediated DNMT1 depletion was instrumental in the acquisition of CSC-like properties [47]. Fan et al. demonstrated that SALL4, an embryonic stem cell transcriptional regulator, is highly expressed in HCC cells and is negatively correlated with HCC prognosis. They also observed significant downstream demethylation of the SALL4 transcriptional start site [48]. The expression of CD133, a CSC marker, is not only associated with poor HCC prognosis, but is also regulated by DNA methylation [49]. Wang et al. showed that hypomethylation of the NANOG promoter and subsequent upregulation are closely associated with p53 depletion. NANOG overexpression results in stem cell-like properties, and promotes HCC development and progression [50]. A recent study found that hypoxia enhances the stemness of HCC cells by enhancing HIF-1α deSUMOylation by SENP1 and increasing the transcriptional activity of HIF-1α, which in turn facilitates HCC stem cell participation in abnormal liver regeneration [51]. Members of the HIF family are highly expressed in the hypoxic microenvironment and activate the c-met and EMT signaling pathways to regulate HCC proliferation, invasion, and metastasis [52–54]. In addition, hypomethylation of hexokinase 2 in HCC cells promotes the interaction between HIF-1α and hypoxia-response element at the −234/−230 site and accelerates HCC progression [55]. The hypoxic microenvironment of HCC also promotes HIF expression and induces DNA demethylation, which subsequently upregulates MAT2A and promotes HCC proliferation and metastasis [56]. Li et al. showed that changes in the DNA methylation level in CSCs suppressed their functions. They found that caffeic acid promoted DNA methylation in the CSCs, which enhanced miR-148a expression, downregulated Samd2, and inhibited HCC progression [57]. Arsenic trioxide can activate microrna-148a by inducing DNA demethylation, thus inhibiting the NF-κB pathway through 3′-UTR targeting p65, making MDR BEL-7402 cells again sensitive to chemotherapy drugs, thus inhibiting the CSC-like phenotype [58]. As more studies focus on the role of DNA methylation in HCC, modulation of stem cell differentiation will become one of the major focuses in HCC research (Figure 1).
Figure 1.
DNA methylation and abnormal liver-regenerating microenvironment in hepatocellular carcinoma.
DNA methylation and HCC
Recent evidence shows that DNA methylation can inhibit gene expression, while abnormal DNA methylation in HCC can lead to inhibition involving cell cycle regulation, faulty DNA repair, and cell apoptosis, as well as inactivation of some tumor-suppressor genes involved in HCC development [59]. EHMT2 (euchromatic histone-lysine N-methyltransferase 2) is a histone methyltransferase, which is mainly responsible for the demethylation of histone h3lysine 9 (H3K9). EHMT2 has been identified as a key mediator in the pathogenesis of HCC. It can inhibit EHMT2, reduce H3K9 methylation, and resist the invasion of liver cancer cells [60]. It has been reported that ACAD (acyl-CoA dehydrogenase) is involved in the proliferation and metastasis of HCC, and DNA methylation plays a key role in regulating the expression of ACADS. A study using knockdown methylation found that the expression of ACADS in HCC cells increased significantly [61]. HBV infection can induce DNA methylation in liver cancer. P16, RASSF1A, GSTP1, APC, p15, and SFRP1 genes in HBV-positive cancer tissues show significant hypermethylation, and DNA methylation can lead to the development of HBV-related HCC [62].
Traditional Chinese Medicine (TCM) and Abnormal LRM
Many new advances in TCM have been made in blocking the occurrence and development of HCC by influencing the abnormal LRM.
Fuzheng Huayu capsule combined with anti-hepatitis B virus (HBV) replication drugs can improve the abnormal LRM (liver fibrosis) significantly better than single-nucleoside antiviral drugs [63,64]. Anluohuaxian capsule in combination with adefovir dipivoxil (ADV) for the treatment of chronic hepatitis B (CHB) patients can improve the abnormal LRM and block the progression of hepatic fibrosis [65]. A component of Radix Bupleuri (Chaihu), saikosaponin-d, attenuated development of liver fibrosis in an animal model. Moreover, saikosaponin-d can inhibit proliferation and transdifferentiation of hepatic stellate cells [66]. Huqi San improves the abnormal LRM by regulating the expression of c-jun, c-fos, and c-Myc, and its efficacy in blocking/reversing precancerous lesions of HCC has been confirmed [67]. Yanggan Jiedu Sanjie inhibits TGF-β1-induced EMT and cell adhesion, migration, and invasion in Bel-7402 cells, and downregulates Smad3 phosphorylation and Snail expression, thus improving the abnormal LRM [68]. Brucea javanica not only reduced the expression of stem cell markers but also eliminated tumor spheroids by apoptotic cell death [69]. Scutellaria Barbate has anti-tumor effects and can eliminate local tumor nodules, promotes blood circulation, regulates the integrity of body function and the interaction between various organs and the internal environment, improves LRM abnormalities, and is widely used in the treatment of various cancers such as liver cancer, lung cancer, stomach cancer, breast cancer, and colorectal cancer [70]. Some studies have shown that Astragalus membranaceus and other herbs for strengthening healthy Qi can regulate human immunity and affect cancer cell cycle, while Coptis chinensis, which has heat-clearing and detoxifying effects, can regulate human immunity and induce cancer cell apoptosis [71,72]. It was found that Kanglaite can treat hepatocellular carcinoma in rats. After intratumoral injection of Kanglaite, the liver function of rats was significantly reduced compared with saline injection, and the inhibition rate was 49.4%. The mechanism of Kanglaite ‘s action may be that it inhibits the expression of proliferating cell nuclear antigen (PCNA) and thus inhibits the proliferation and mitosis of cancer cells. Kanglaite also induces apoptosis of HepG2 cells by activating the fas/fasl pathway, and improves abnormal LRM [73,74].
The TCM formula QHF (Q, Qingrejiedu; H, Huoxuehuayu; and F, Fuzhengguben) inhibits the proliferation of HepG2 cells and decreases their ability to invade and metastasize [75]. The anti-cachexia effects of jianpijiedu (MJPJD) appear to be associated with the downregulation of the expression levels of the inflammatory cytokines IL-1α, IL-6, and TNF-α. MJPJD exerts effects at the molecular levels, such as affecting the expression of cytokines MURF1 and atrogin 1, and also affects ascites, body weight, maintenance of net weight, body temperature, and food intake, and can act against abnormal LRM [76]. Diwu Yanggan capsule can alleviate the development of liver fibrosis by improving the abnormal LRM through regulating the EMT/MET imbalance (inhibiting EMT and promoting MET) [77]. Further studies showed that Diwu Yanggan capsule can inhibit over-activation of the Wnt/beta-catenin pathway, inhibit the over-proliferation and abnormal differentiation of oval cells, and prevent the occurrence and development of precancerous lesions of HCC [78]. A randomized controlled clinical trial showed that Diwu Yanggan capsule improves the hepatic histological response in patients with HBeAg-negative chronic hepatitis B, which significantly reduced the occurrence of cirrhosis, improved the abnormal LRM, and then reduced the incidence of HCC risk [79]. In recent years it is found that many kinds of Chinese medicines can inhibit angiogenesis, decrease the liver cancer microvessel density, and improve the abnormal LRM, thus delaying or preventing the development of liver cancer [80].
TCM and DNA Methylation
The influence of DNA methylation is one of the mechanisms through which TCM plays a functional role. TCM syndrome types are linked to related gene epigenetic modifications, and this may be one of the molecular mechanisms underlying the occurrence, development, and differentiation. Phlegm-dampness constitution (PC) is one of the various constitution types in TCM, which is considered to be the preclinical stage of various metabolic disorders. Data of the genomic DNA methylation profile in human peripheral blood mononuclear cells of PC and balanced constitution (BC) subjects suggests that DNA methylation can play a role in the pathogenesis of metabolic diseases during the incubation period of HCC [81]. There is a unique methylation level change of the synovium gene in the model of hot arthralgia, and Baihu jiaguizhi decoction can specifically reduce the methylation level of the characteristic gene of hot arthralgia, so as to achieve the effect of treating hot arthralgia [82]. CHM compounds were able to change DNA methylation levels. Curcumin, which has diverse functions, can promote corresponding gene expression via down-regulating DNA gene methylation in blood cancer [83–85]. Jieduquyuziyin regulates immune function by increasing the level of genomic DNA methylation and MeCP2 gene promoter region methylation in Jurkat cells [86]. In breast cancer, the expression of the ras-association domain family protein 1A is reactivated by curcumin through decreasing methylation of its promoter [87]. Berberine also can induce apoptosis in the human multiple myeloma cell line U266 through hypomethylation of p53 promoter [88]. Resveratrol has been shown to affect DNA methylation, and in many cancer cell lines the expression of DNMT1, DNMT3A, and DNMT3B were downregulated by resveratrol [89]. Quercetin can arrest human RKO colon cancer cell cycle by demethylating the CDKN2A promoter [90]. Dietary polyphenol quercetin or butyrate can reverse epigenetic changes by decreasing DNA methylation in cancer cells and can be used as chemopreventive and therapeutic drugs for cancer [91]. Parthenolide inhibits MCF-7 tumor cell growth by increasing the expression of HIN-1 through regulating the hypomethylation of HIN-1 promoter [92]. Chlorogenic acid can inhibit DNA methylation in the promoter of tumor-suppressor gene RARB in the cancer cell line MCF-7 [93]. Epigallocatechin-3-gallate (EGCG, found in green tea) reduced the overall DNA methylation level of A431 cells, and treated the re-expression of the silent oncogene p16INK4a and ip1/p21, which may contribute to the chemoprevention of skin cancer and provide new insights into the epigenetic mechanism [94]. Typical epigenetic dietary agents such as resveratrol isoflavones sulforaphane and garlic compounds in dietary fiber have been reported to correct and stabilize epigenetic modification [95–97]. The arsenic-containing Chinese herbal formulae (CHF) Qinghuang causes significant genome-wide demethylation, and affects chromosome karyotype status and genome methylation level [98]. Dujieqing oral liquid can achieve effective synergy and reduce toxicity by regulating gene methylation activity, targeting patients with advanced cancer in chemotherapy [99]. In addition, further studies have shown that the regulation of human epigenome by CHF is helpful to elucidate the synergistic effects of H3K9 methylation and H3K4 demethylation on heterochromatinization in the discovery of plant pharmacology and epigenetic drugs. Chinese herbal compounds mainly inhibit DNA methyltransferase and histone deacetylase by synergistically inhibiting histone acetylation and H3S10 phosphorylation or H3K4 demethylation and H3K36 demethylation [96]. Ethanol extract of Ligustrum lucidum Ait leaves (EEl) can downregulate the DNA methylation level of PTEN and increase the expression of unmethylated PTEN, helping to inhibit the PI3K/Akt pathway, regulate the expression of factors related to apoptosis cell cycle arrest (Bax, Bcl-2, cytc, Caspase-3, Ki67, CyclinD1 and p21), thus inducing apoptosis and promoting cell cycle arrest, and reducing the invasion and migration of HCC cells [100]. Research shows that Chuanxiong capsule can regulate the abnormal hypermethylation and hypomethylation genes in an atherosclerosis rabbit model and participate in the activity of protein kinase C activity, inflammatory pathway, MAPK signaling pathway, and VEGF signaling pathway [101]. Recently, telomere and telomerase have been studied as targets of anticancer drugs. Studies have shown that telomerase reverse transcriptase is a potential target gene involved in the effect of the Chinese herbal medicine Tianshengyuan-1 (Tsy-1). It can increase telomerase activity in normal peripheral blood monocytes and CD34+ hematopoietic stem cells with low telomerase activity, and the effect is related to the methylation of tert promoter [102]. However, there has been little research on the relationship between TCM and DNA demethylation. Considering the diverse curative effects and minimal adverse effects of TCM, the mechanisms, especially the regulation mechanism of DNA methylation by TCM, require further investigation.
Concluding Remarks
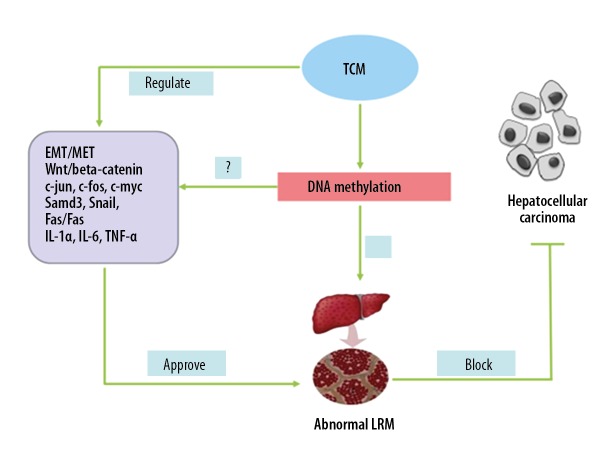
DNA methylation is a critical epigenetic mechanism regulating HCC development and progression, as well as the LRM. Altering DNA methylation levels and patterns and improving the LRM are potential therapeutic measures against HCC. However, drugs targeting DNA methylation have not been extensively tested. Single-target drugs are often ineffective due to the multiple methylation sites throughout the genome, multiple methylation patterns, and inability to balance genomic hypermethylation and demethylation. In addition, lack of evidence of a direct association between abnormal single-site methylation and HCC prognosis further limits the development of methylation-targeting drugs. Traditional Chinese medicine (TCM) is increasingly being considered as a therapeutic intervention targeting DNA methylation due to its multi-component, multi-targeting, and multi-action formulations. TCM has shown promising results in HCC prevention and treatment via LRM improvement. However, their mechanisms of action need further validation [17]. In addition, further understanding of the mechanisms underlying abnormal DNA methylation and LRM in HCC and improvement of LRM via overall dynamic and targeted regulation of DNA methylation will undoubtedly be the future research directions in HCC (Figure 2).
Figure 2.
TCM between DNA methylation and abnormal liver-regenerating microenvironment.
Footnotes
Source of support: This study was financially supported by the National Natural Science Foundation of China (No. 81373513, No. 90709041, No. 306725-90, No. 30271562, No. 30371787, No. 8110-2531, No. 81274147, No. 81373513, No. 81-573815, No. 81603484, and No. 81703912), the National TCM Clinical Project (JDZX2012-054 and JDZX2015172), Natural Science Foundation of Hubei Province (No. 2017CFB380), the National Nonprofit Institute Research Grant for Institute of Basic Theory for Chinese Medicine, CACMS (YZ-1615) and Hanmin Li National Famous Old Chinese Medicine Experts Inheritance Studio, Guangxi Liver Disease Clinical Research Center of Traditional Chinese Medicine, and the National Clinical Research Base of Traditional Chinese Medicine (Guangxi Province), the National Natural Science Foundation of China (No. 81774236, 81704021, No. 81760844), Guangxi Natural Science Foundation (2017GXNSFBA198200, 2018GXNSFAA281047), Innovation research team project of Guangxi Natural Science Foundation (2018GXNSFGA281002)
Conflict of interest
None.
References
- 1.Chaturvedi VK, Singh A, Dubey SK, et al. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb Pathog. 2019;128:184–94. doi: 10.1016/j.micpath.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Yin T, Xu Y, Lu XJ. Therapeutics for advanced hepatocellular carcinoma: Recent advances, current dilemma, and future directions. J Cell Physiol. 2019;234(8):12122–32. doi: 10.1002/jcp.28048. [DOI] [PubMed] [Google Scholar]
- 3.Wallace MC, Preen D, Jeffrey GP, Adams LA. The evolving epidemiology of hepatocellular carcinoma: A global perspective. Exp Rev Gastroenterol Hepatol. 2015;9(6):765–79. doi: 10.1586/17474124.2015.1028363. [DOI] [PubMed] [Google Scholar]
- 4.Li HM. Microcirculation of liver cancer, microenvironment of liver regeneration, and the strategy of Chinese medicine. Chin J Integr Med. 2016;22(3):163–67. doi: 10.1007/s11655-016-2460-y. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Zhang L. Liver regeneration microenvironment of hepatocellular carcinoma for prevention and therapy. Oncotarget. 2017;8(1):1805–13. doi: 10.18632/oncotarget.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura M, Chiba T, Kanayama K, et al. Epigenetic dysregulation in hepatocellular carcinoma: an up-to-date review. Hepatol Res. 2019;49(1):3–13. doi: 10.1111/hepr.13250. [DOI] [PubMed] [Google Scholar]
- 7.Corella D, Ordovas JM. Basic concepts in molecular biology related to genetics and epigenetics. Rev Esp Cardiol (Engl Ed) 2017;70(9):744–53. doi: 10.1016/j.rec.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pishva E, Rutten BPF, van den Hove D. DNA methylation in major depressive disorder. Adv Exp Med Biol. 2017;978:185–96. doi: 10.1007/978-3-319-53889-1_10. [DOI] [PubMed] [Google Scholar]
- 10.Tahmasebi Birgani M, Carloni V. Tumor microenvironment, a paradigm in hepatocellular carcinoma progression and therapy. Int J Mol Sci. 2017;18(2) doi: 10.3390/ijms18020405. pii: E405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Chen L. Tumor microenviroment and hepatocellular carcinoma metastasis. J Gastroenterol Hepatol. 2013;28(Suppl 1):43–48. doi: 10.1111/jgh.12091. [DOI] [PubMed] [Google Scholar]
- 12.Li HM. [Research progress and prospect of Traditional Chinese Medicine in regulating liver regeneration]. World Chinese Journal of Digestology. 2017;25(15):1338–44. [in Chinese] [Google Scholar]
- 13.Li HM, Ye ZH. Microenvironment of liver regeneration in liver cancer. Chin J Integr Med. 2017;23(7):555–60. doi: 10.1007/s11655-017-2806-0. [DOI] [PubMed] [Google Scholar]
- 14.Wallace MC, Friedman SL. Hepatic fibrosis and the microenvironment: Fertile soil for hepatocellular carcinoma development. Gene Expr. 2014;16(2):77–84. doi: 10.3727/105221614X13919976902057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morse MA, Sun W, Kim R, et al. The role of angiogenesis in hepatocellular carcinoma. Clin Cancer Res. 2019;25(3):912–20. doi: 10.1158/1078-0432.CCR-18-1254. [DOI] [PubMed] [Google Scholar]
- 16.Critelli R, Milosa F, Faillaci F, et al. Microenvironment inflammatory infiltrate drives growth speed and outcome of hepatocellular carcinoma: A prospective clinical study. Cell Death Dis. 2017;8(8):e3017. doi: 10.1038/cddis.2017.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang N, Wang S, Li MY, et al. Cancer stem cells in hepatocellular carcinoma: An overview and promising therapeutic strategies. Ther Adv Med Oncol. 2018;10 doi: 10.1177/1758835918816287. 1758835918816287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li HM. [Progress and prospect of liver regeneration microenvironment in hepatocellular carcinoma (review)]. World Chinese Journal of Digestology. 2018;26(26):1529–36. [in Chinese] [Google Scholar]
- 19.Ni S, Li S, Yang N, et al. Deregulation of regulatory T cells in acute-on-chronic liver failure: A rat model. Mediators Inflamm. 2017;2017 doi: 10.1155/2017/1390458. 1390458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li HM. [The basis and clinic of TCM regulating liver regeneration]. Huazhong University of Science and Technology Press; 2016. [in Chinese] [Google Scholar]
- 21.Li HMZ, Gao X. [Effect and mechanism of “invigorating kidney, generating marrow and forming liver” on improving microenvironment of liver regeneration in preventing and treating hepatocellular carcinoma]. Journal of Hubei University of Traditional Chinese Medicine. 2015;17(1):5–8. [in Chinese] [Google Scholar]
- 22.Li HM. [The necessity and contingency of TCM research are unified in probabilities and probabilities]. Chinese Archives of Traditional Chinese Medicine. 2018;36(8):1799–802. [in Chinese] [Google Scholar]
- 23.Schrader J, Gordon-Walker TT, Aucott RL, et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology (Baltimore, Md) 2011;53(4):1192–205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann J, Oakley F, Akiboye F, et al. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: Implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14(2):275–85. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- 25.Yang JJ, Tao H, Huang C, et al. DNA methylation and MeCP2 regulation of PTCH1 expression during rats hepatic fibrosis. Cell Signal. 2013;25(5):1202–11. doi: 10.1016/j.cellsig.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Reister S, Kordes C, Sawitza I, Haussinger D. The epigenetic regulation of stem cell factors in hepatic stellate cells. Stem Cells Dev. 2011;20(10):1687–99. doi: 10.1089/scd.2010.0418. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Wu XQ, Li WX, et al. PSTPIP2 connects DNA methylation to macrophage polarization in CCL4-induced mouse model of hepatic fibrosis. Oncogene. 2018;37(47):6119–35. doi: 10.1038/s41388-018-0383-0. [DOI] [PubMed] [Google Scholar]
- 28.Ogata H, Chinen T, Yoshida T, et al. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene. 2006;25(17):2520–30. doi: 10.1038/sj.onc.1209281. [DOI] [PubMed] [Google Scholar]
- 29.Folkman J. Fundamental concepts of the angiogenic process. Curr Mol Med. 2003;3(7):643–51. doi: 10.2174/1566524033479465. [DOI] [PubMed] [Google Scholar]
- 30.Ridge SM, Sullivan FJ, Glynn SA. Mesenchymal stem cells: Key players in cancer progression. Mol Cancer. 2017;16(1):31. doi: 10.1186/s12943-017-0597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis. 2017;20(4):409–26. doi: 10.1007/s10456-017-9562-9. [DOI] [PubMed] [Google Scholar]
- 32.Ge H, Luo H. Overview of advances in vasculogenic mimicry – a potential target for tumor therapy. Cancer Manag Res. 2018;10:2429–37. doi: 10.2147/CMAR.S164675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips JM, Goodman JI. Identification of genes that may play critical roles in phenobarbital (PB)-induced liver tumorigenesis due to altered DNA methylation. Toxicol Sci. 2008;104(1):86–99. doi: 10.1093/toxsci/kfn063. [DOI] [PubMed] [Google Scholar]
- 34.Phillips JM, Goodman JI. Multiple genes exhibit phenobarbital-induced constitutive active/androstane receptor-mediated DNA methylation changes during liver tumorigenesis and in liver tumors. Toxicol Sci. 2009;108(2):273–89. doi: 10.1093/toxsci/kfp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quentmeier H, Eberth S, Romani J, et al. DNA methylation regulates expression of VEGF-R2 (KDR) and VEGF-R3 (FLT4) BMC Cancer. 2012;12:19. doi: 10.1186/1471-2407-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li GJ, Harrison TJ, Yang JY, et al. Combined core promoter mutations and pre-S deletion of HBV may not increase the risk of HCC: A geographical epidemiological study in Guangxi, China. Liver Int. 2013;33(6):936–43. doi: 10.1111/liv.12142. [DOI] [PubMed] [Google Scholar]
- 37.Fu J, Xu D, Liu Z, et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology. 2007;132(7):2328–39. doi: 10.1053/j.gastro.2007.03.102. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y, Cheng Y, Yan W, Nardini C. Exploring the molecular causes of hepatitis B virus vaccination response: An approach with epigenomic and transcriptomic data. BMC Med Genomics. 2014;7:12. doi: 10.1186/1755-8794-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto Y, Shinjo K, Shimizu Y, et al. Hepatitis virus infection affects DNA methylation in mice with humanized livers. Gastroenterology. 2014;146(2):562–72. doi: 10.1053/j.gastro.2013.10.056. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Petropoulos S, Liu J, et al. The signature of liver cancer in immune cells DNA methylation. Clin Epigenetics. 2018;10:8. doi: 10.1186/s13148-017-0436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jelencic V, Lenartic M, Wensveen FM, Polic B. NKG2D: A versatile player in the immune system. Immunol Lett. 2017;189:48–53. doi: 10.1016/j.imlet.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Sanchez A, Baragano Raneros A, Carvajal Palao R, et al. DNA demethylation and histone H3K9 acetylation determine the active transcription of the NKG2D gene in human CD8+ T and NK cells. Epigenetics. 2013;8(1):66–78. doi: 10.4161/epi.23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Wei H, Sun R, et al. Increased susceptibility to liver injury in hepatitis B virus transgenic mice involves NKG2D-ligand interaction and natural killer cells. Hepatology (Baltimore, Md) 2007;46(3):706–15. doi: 10.1002/hep.21872. [DOI] [PubMed] [Google Scholar]
- 44.Flores-Tellez TN, Villa-Trevino S, Pina-Vazquez C. Road to stemness in hepatocellular carcinoma. World J Gastroenterol. 2017;23(37):6750–76. doi: 10.3748/wjg.v23.i37.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li GC, Ye QH, Xue YH, et al. Human mesenchymal stem cells inhibit metastasis of a hepatocellular carcinoma model using the MHCC97-H cell line. Cancer Sci. 2010;101(12):2546–53. doi: 10.1111/j.1349-7006.2010.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuzillet C, de Gramont A, Tijeras-Raballand A, et al. Perspectives of TGF-beta inhibition in pancreatic and hepatocellular carcinomas. Oncotarget. 2014;5(1):78–94. doi: 10.18632/oncotarget.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raggi C, Factor VM, Seo D, et al. Epigenetic reprogramming modulates malignant properties of human liver cancer. Hepatology (Baltimore, Md) 2014;59(6):2251–62. doi: 10.1002/hep.27026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan H, Cui Z, Zhang H, et al. DNA demethylation induces SALL4 gene re-expression in subgroups of hepatocellular carcinoma associated with Hepatitis B or C virus infection. Oncogene. 2017;36(17):2435–45. doi: 10.1038/onc.2016.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.You H, Ding W, Rountree CB. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-beta. Hepatology (Baltimore, Md) 2010;51(5):1635–44. doi: 10.1002/hep.23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang XQ, Ng RK, Ming X, et al. Epigenetic regulation of pluripotent genes mediates stem cell features in human hepatocellular carcinoma and cancer cell lines. PLoS One. 2013;8(9):e72435. doi: 10.1371/journal.pone.0072435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui CP, Wong CC, Kai AK, et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1alpha deSUMOylation and SENP1/HIF-1alpha positive feedback loop. Gut. 2017;66(12):2149–59. doi: 10.1136/gutjnl-2016-313264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu P, Fang X, Song Y, et al. Expression of hypoxia-inducible factor 3alpha in hepatocellular carcinoma and its association with other hypoxia-inducible factors. Exp Ther Med. 2016;11(6):2470–76. doi: 10.3892/etm.2016.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu C, Yang SL, Fang X, et al. Hypoxia disrupts the expression levels of circadian rhythm genes in hepatocellular carcinoma. Mol Med Rep. 2015;11(5):4002–8. doi: 10.3892/mmr.2015.3199. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Li H, Huang Z, et al. Hypoxia attenuates Hsp90 inhibitor 17-DMAG-induced cyclin B1 accumulation in hepatocellular carcinoma cells. Cell Stress Chaperones. 2016;21(2):339–48. doi: 10.1007/s12192-015-0664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee HG, Kim H, Son T, et al. Regulation of HK2 expression through alterations in CpG methylation of the HK2 promoter during progression of hepatocellular carcinoma. Oncotarget. 2016;7(27):41798–810. doi: 10.18632/oncotarget.9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Q, Liu L, Zhao Y, et al. Hypoxia induces genomic DNA demethylation through the activation of HIF-1alpha and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10(6):1113–23. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Jiang F, Chen L, et al. Blockage of TGFbeta-SMAD2 by demethylation-activated miR-148a is involved in caffeic acid-induced inhibition of cancer stem cell-like properties in vitro and in vivo. FEBS Open Bio. 2015;5:466–75. doi: 10.1016/j.fob.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Jiang F, Jiao K, et al. De-methylation of miR-148a by arsenic trioxide enhances sensitivity to chemotherapy via inhibiting the NF-kappaB pathway and CSC like properties. Exp Cell Res. 2019;386(2):111739. doi: 10.1016/j.yexcr.2019.111739. [DOI] [PubMed] [Google Scholar]
- 59.Schagdarsurengin U, Wilkens L, Steinemann D, et al. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22(12):1866–71. doi: 10.1038/sj.onc.1206338. [DOI] [PubMed] [Google Scholar]
- 60.Gu M, Toh TB. Nanodiamond-mediated delivery of a G9a inhibitor for hepatocellular carcinoma therapy. ACS Appl Mater Interfaces. 2019;11(49):45427–41. doi: 10.1021/acsami.9b16323. [DOI] [PubMed] [Google Scholar]
- 61.Chen D, Feng X, Lv Z, et al. ACADS acts as a potential methylation biomarker associated with the proliferation and metastasis of hepatocellular carcinomas. Aging. 2019;11(20):8825–44. doi: 10.18632/aging.102292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C, Huang C, Sui X, et al. Association between gene methylation and HBV infection in hepatocellular carcinoma: A meta-analysis. J Cancer. 2019;10(25):6457–65. doi: 10.7150/jca.33005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu P, Hu YY, Liu C, et al. Multicenter clinical study on Fuzhenghuayu capsule against liver fibrosis due to chronic hepatitis B. World J Gastroenterol. 2005;11(19):2892–99. doi: 10.3748/wjg.v11.i19.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu P. Fuzheng huayu capsule in the treatment of liver fibrosis: Clinical evidence and mechanism of action. Chin J Integr Med. 2012;18(5):398–400. doi: 10.1007/s11655-012-1030-1. [DOI] [PubMed] [Google Scholar]
- 65.Jiang YF, Ma J, He B, et al. [The therapeutic effect of Anluohuaxian capsule combined with adefovir dipivoxil on patients with chronic hepatitis B and influence on hepatic histology]. Zhonghua Gan Zang Bing Za Zhi. 2012;20(5):344–47. doi: 10.3760/cma.j.issn.1007-3418.2012.05.008. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 66.Gong Y. Identifying the targets for treatment of liver fibrosis and hepatocellular carcinoma from both Western medicine and Chinese medicine. Chin J Integr Med. 2012;18(4):245–49. doi: 10.1007/s11655-012-1062-6. [DOI] [PubMed] [Google Scholar]
- 67.Li X, Shi ZM, Feng P, et al. Effect of Qi-protecting powder (Huqi San) on expression of c-jun, c-fos and c-myc in diethylnitrosamine-mediated hepatocarcinogenesis. World J Gastroenterol. 2007;13(31):4192–98. doi: 10.3748/wjg.v13.i31.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu B, An HM, Yan X, et al. Traditional Chinese medicine formulation Yanggan Jiedu Sanjie inhibits TGF-beta1-induced epithelial-mesenchymal transition and metastatic potential in human hepatocarcinoma Bel-7402 cells. BMC Complement Altern Med. 2019;19(1):67. doi: 10.1186/s12906-019-2477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen JH, Kim SH, Fan PW, et al. The aqueous extract of Chinese medicinal herb Brucea javanica suppresses the growth of human liver cancer and the derived stem-like cells by apoptosis. Drug Des Devel Ther. 2016;10:2003–13. doi: 10.2147/DDDT.S107909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng Z, Cho WC, Xu L, et al. Lessons learnt from evidence-based approach of using chinese herbal medicines in liver cancer. Evid Based Complement Alternat Med. 2013;2013 doi: 10.1155/2013/656351. 656351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu GL, Zhang L, Li TY, et al. Short-term effect of combined therapy with Jinlong Capsule and transcatheter arterial chemoembolization on patients with primary hepatic carcinoma and its influence on serum osteopontin expression. Chinese J Integr Med. 2010;16(2):109–13. doi: 10.1007/s11655-010-0109-9. [DOI] [PubMed] [Google Scholar]
- 72.Yong S, Da Z, et al. [HPLC separation and determination of bufadienolide in cinobufacini injection]. Chinese Traditional Patent Medicine. 2003;24(1):4. [in Chinese] [Google Scholar]
- 73.Wu LQ, Lu Y, Lu HJ, et al. Efficacy of intra-tumor injection of Kang-Lai-Te in treating transplanted hepatoma in rats. Hepatobiliary Pancreat Dis Int. 2004;3(4):580–84. [PubMed] [Google Scholar]
- 74.Lu Y, Wu LQ, Dong Q, Li CS. Experimental study on the effect of Kang-Lai-Te induced apoptosis of human hepatoma carcinoma cell HepG2. Hepatobiliary Pancreat Dis Int. 2009;8(3):267–72. [PubMed] [Google Scholar]
- 75.Chen T, Wang Q, Li Y, et al. Chinese herbal formula QHF inhibits liver cancer cell invasion and migration. Exp Ther Med. 2016;11(6):2413–19. doi: 10.3892/etm.2016.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun B, Luo H, Deng L, et al. The study on mechanism of the modified Chinese herbal compound, jianpijiedu, on a mouse model of hepatic carcinoma cachexia. Mol Med Rep. 2016;14(4):3113–21. doi: 10.3892/mmr.2016.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen X, Cheng S, Peng Y, et al. Attenuation of early liver fibrosis by herbal compound “Diwu Yanggan” through modulating the balance between epithelial-to-mesenchymal transition and mesenchymal-to-epithelial transition. BMC Complement Altern Med. 2014;14:418. doi: 10.1186/1472-6882-14-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao BB, Li HM, Gao X, et al. The herbal compound “diwu yanggan” modulates liver regeneration by affecting the hepatic stem cell microenvironment in 2-acetylaminofluorene/partial hepatectomy rats. Evid Based Complement Alternat Med. 2015;2015 doi: 10.1155/2015/468303. 468303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H, Ye Z, Gao X, et al. Diwu Yanggan capsule improving liver histological response for patients with HBeAg-negative chronic hepatitis B: A randomized controlled clinical trial. Am J Transl Res. 2018;10(5):1511–21. [PMC free article] [PubMed] [Google Scholar]
- 80.LX . Journal of Modern Oncology. 2014. [in Chinese] [Google Scholar]
- 81.Yao H, Mo S, Wang J, et al. Genome-wide DNA methylation profiles of phlegm-dampness constitution. Cell Physiol Biochem. 2018;45(5):1999–2008. doi: 10.1159/000487976. [DOI] [PubMed] [Google Scholar]
- 82.Chen H, Ju SH, Wei JP, et al. [Effect of Baihu Guizhi decoction on characteristic methylation genes expression of pyretic arthralgia rat model]. Zhongguo Zhong Yao Za Zhi. 2017;42(2):332–40. doi: 10.19540/j.cnki.cjcmm.20161222.013. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 83.Yu J, Peng Y, Wu LC, et al. Curcumin down-regulates DNA methyltransferase 1 and plays an anti-leukemic role in acute myeloid leukemia. PLoS One. 2013;8(2):e55934. doi: 10.1371/journal.pone.0055934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu XM, Shen JZ, Shen SF, Fan LP. [Effect of triptolide on reversing hypermethylation of apc gene in Jurkat cells and its possible mechanisms]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18(4):866–72. [in Chinese] [PubMed] [Google Scholar]
- 85.Papoutsis AJ, Lamore SD, Wondrak GT, et al. Resveratrol prevents epigenetic silencing of BRCA-1 by the aromatic hydrocarbon receptor in human breast cancer cells. J Nutr. 2010;140(9):1607–14. doi: 10.3945/jn.110.123422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li R, Zhuang A, Ma J, et al. Effect of Jieduquyuziyin prescription-treated rat serum on MeCP2 gene expression in Jurkat T cells. Cancer Med. 2018;54(10):692–704. doi: 10.1007/s11626-018-0295-x. [DOI] [PubMed] [Google Scholar]
- 87.Dammann RH, Richter AM, Jimenez AP, et al. Impact of natural compounds on DNA methylation levels of the tumor suppressor gene RASSF1A in cancer. Int J Mol Sci. 2017;18(10) doi: 10.3390/ijms18102160. pii: E2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qing Y, Hu H, Liu Y, et al. Berberine induces apoptosis in human multiple myeloma cell line U266 through hypomethylation of p53 promoter. Cell Biol Int. 2014;38(5):563–70. doi: 10.1002/cbin.10206. [DOI] [PubMed] [Google Scholar]
- 89.Fernandes GFS, Silva GDB, Pavan AR, et al. Epigenetic regulatory mechanisms induced by resveratrol. Nutrients. 2017 Nov 1;9(11) doi: 10.3390/nu9111201. pii: E1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tan S, Wang C, Lu C, et al. Quercetin is able to demethylate the p16INK4a gene promoter. Chemotherapy. 2009;55(1):6–10. doi: 10.1159/000166383. [DOI] [PubMed] [Google Scholar]
- 91.Zheng NG, Wang JL, Yang SL, Wu JL. Aberrant epigenetic alteration in Eca9706 cells modulated by nanoliposomal quercetin combined with butyrate mediated via epigenetic-NF-kappaB signaling. Asian Pac J Cancer Prev. 2014;15(11):4539–43. doi: 10.7314/apjcp.2014.15.11.4539. [DOI] [PubMed] [Google Scholar]
- 92.Liu Z, Liu S, Xie Z, et al. Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J Pharmacol Exp Ther. 2009;329(2):505–14. doi: 10.1124/jpet.108.147934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dai X, Huang Q, Zhou B, et al. Preparative isolation and purification of seven main antioxidants from Eucommia ulmoides Oliv. (Du-zhong) leaves using HSCCC guided by DPPH-HPLC experiment. Food Chem. 2013;139(1–4):563–70. doi: 10.1016/j.foodchem.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 94.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32(4):537–44. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang Y, Song H, Hu H, et al. Trichosanthin inhibits DNA methyltransferase and restores methylation-silenced gene expression in human cervical cancer cells. Mol Med Rep. 2012;6(4):872–78. doi: 10.3892/mmr.2012.994. [DOI] [PubMed] [Google Scholar]
- 96.Hsieh HY, Chiu PH, Wang SC. Histone modifications and traditional Chinese medicinals. BMC Complement Altern Med. 2013;13:115. doi: 10.1186/1472-6882-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim SH, Bommareddy A, Singh SV. Garlic constituent diallyl trisulfide suppresses x-linked inhibitor of apoptosis protein in prostate cancer cells in culture and in vivo. Cancer Prev Res (Phila) 2011;4(6):897–906. doi: 10.1158/1940-6207.CAPR-10-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shuzhen S, Rou M, Xiaomei H, et al. Karyotype and DNA-methylation responses in myelodysplastic syndromes following treatment with traditional chinese formula containing arsenic. Evid Based Complement Alternat Med. 2012;2012 doi: 10.1155/2012/969476. 969476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rong Z, Xu Y, Mo CM. [Effects of dujieqing oral liquid on the promoter methylation of the MGMT gene in middle-and-late stage tumor patients receiving chemotherapy]. Zhongguo Zhong Xi Yi Jie He Za. 2012;32(12):1611–15. [PubMed] [Google Scholar]
- 100.Tian G, Chen J, Luo Y, et al. Ethanol extract of Ligustrum lucidum Ait. leaves suppressed hepatocellular carcinoma in vitro and in vivo. 2019;19:246. doi: 10.1186/s12935-019-0960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou QB, Wu LQ, Zhang Y, et al. Effects of Zhizi Chuanxiong Capsule () on the Abnormal Methylation in Rabbits with Atherosclerosis. Chin J Integ Med. 2018;24(7):512–17. doi: 10.1007/s11655-018-2561-x. [DOI] [PubMed] [Google Scholar]
- 102.Yu W, Qin X, Jin Y, et al. Tianshengyuan-1 (TSY-1) regulates cellular telomerase activity by methylation of TERT promoter. Oncotarget. 2017;8(5):7977–88. doi: 10.18632/oncotarget.13939. [DOI] [PMC free article] [PubMed] [Google Scholar]