Abstract
The associations of four proteins – AK4, ITPK1, HSPB2, and IGFBP5 – with cognitive function in older adults were largely unexplained by known brain pathologies. We examined the extent to which individual protein associations with cognitive decline were attributable to microstructural changes in the brain. This study included 521 participants (mean age 90.3, 65.9–108.3) with the postmortem reciprocal of transverse relaxation time (R2) magnetic resonance image. All participants came from one of two ongoing longitudinal cohorts of aging and dementia, the Religious Orders Study and Rush Memory and Aging Project.
Higher abundance of AK4, HSPB2, and IGFBP5 was associated with faster cognitive decline, and mediated through lower postmortem R2 in the frontal and temporal white matter regions. In contrast, higher abundance of ITPK1 was associated with slower cognitive decline, and mediated through higher postmortem R2 in the frontal and temporal white matter regions. The association of four proteins - AK4, ITPK1, IGFBP5, and HSPB2 - with cognition in late life were explained via microstructural changes in the brain.
1. Introduction
RNA sequencing data from postmortem human brains showed a robust relationship between cognitive decline – a clinical hallmark of Alzheimer’s dementia – and a network of coexpressed cortical genes (Mostafavi et al., 2018). This coexpressed network is likely related to transcriptional regulation and termed ‘module 109’ (m109). The average expression of m109 was also related to burden of brain β-amyloid deposition – an Alzheimer’s Disease (AD) pathology (Mostafavi et al., 2018). In a subsequent study (Yu et al., 2018), we prioritized 30 genes out of 390 genes involved in the m109 as potential drivers of the module. Out of those 30 genes, 14 genes were tested for their knockdown of gene expression in human astrocytes cell culture derived from induced pluripotent stem cell (iPSC), and measured for its extracellular levels of the pathogenic β-amyloid Aβ42 peptide. Out of 14 genes, inositol polyphosphate phosphatase like 1 (INPPL1) and plexin B1 (PLXNB1) RNA knockdown reproducibly reduced the production of pathogenic soluble Aβ42. These experiments suggested that INPPL1 and PLXNB1 may play an important role of m109 on β-amyloid metabolism. Although m109 was causally implicated in Alzheimer’s dementia in part through brain β-amyloid deposition, it also had a strong effect on cognitive decline that was independent of β-amyloid and age-related pathologies (Yu et al., 2018). Using selected reaction monitoring (SRM) quantitative proteomics it was possible to quantify 12 of the corresponding proteins. We examined their association with AD and non-AD pathology beyond β-amyloid deposition, as well as cognitive decline. Of these, five proteins were associated with cognitive decline. The association of one protein – PLXNB1 – with cognitive decline was mediated primarily through β-amyloid load and PHFtau tangle density. However, the associations of insulin like growth factor binding protein 5 (IGFBP5) and heat shock protein family B member 2 (HSPB2) were only partially mediated by AD or other age-related pathologies – amyolid, PHFtau tangles, macroinfarcts, microinfarcts, lewy bodies, hippocampal sclerosis, TDP stage 4, cerebral amyloid angiopathy, cerebral atherosclerosis, and arteriolsclerosis (Yu et al., 2018). Furthermore, adenylate kinase 4 (AK4) and inositol-tetrakisphosphate 1-kinase (ITPK1) were essentially independent of common brain pathologies (Yu et al., 2018). Interestingly, higher abundance of AK4, HSPB2, and IGFBP5 were associated with faster cognitive decline, whereas higher abundance of ITPK1 was associated with slower cognitive decline (Yu et al., 2018).
Recent combinations of molecular levels and neuroimaging has shown that many features of brain structure and function have analogues on the molecular level (Fulcher and Fornito, 2016, Krienen et al., 2016, Richiardi et al., 2015). These studies typically compare neuroimaging and molecular levels acquired from distinct areas of the brain. Because brains donated by participants from the Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP) undergo postmortem imaging it is possible to map the brain regions that covary with levels of a given molecule. To account for the unexplained covariation of AK4, HSPB2, IGFBP5, and ITPK1 with cognitive decline, we considered the potential for these proteins to play a role in maintaining or degrading microstructure. Therefore, we examined the extent to which brain microstructure as assessed with R2 could mediate the association of these four brain proteins with cognitive decline. Among various postmortem brain imaging modalities, R2, the reciprocal of transverse relaxation time (i.e., T2) on MRI, has been studied for its association with late life cognitive decline and brain pathologies (Arfanakis, Konstantinos et al., 2007, Dawe et al., 2014a, Dawe et al., 2016, Dawe et al., 2018, House et al., 2006, House et al., 2008, Kirsch et al., 1992, Wang et al., 2004). For instance, previous reports have associated in vivo R2 with different stages of memory impairment (House et al., 2006), where lower R2 in widespread white matter regions and higher R2 in the grey matter of the temporal lobe coincided with impaired memory. One potential cause of lower R2, or higher T2, in white matter is increased water content in white matter tissue due to breakdown of myelin by neurodegenerative processes and vascular insults. While biological mechanisms of neurodegeneration in grey matter are more complicated (House et al., 2006), one cause for higher R2 in grey matter may be higher accumulation of iron. Higher iron content is known to cause local distortion of the magnetic field that leads to higher relaxation rate, i.e. shorter T2, and thus higher R2. Due to the relevance of R2 to both brain structure and cognition, we examine how it could mediate the relationship of protein abundance with cognition, particularly in cases when AD or other age-related pathologies do not account for their relationship. We utilize a subset of the same study subjects - ROS and MAP – that were the subject of prior work (Mostafavi et al., 2018, Yu et al., 2018) based on data metrics required for these analyses.
2. Methods
2.1. Participants
Participants were older adults enrolled in one of two ongoing cohort studies of aging and dementia, ROS and MAP (Bennett, D. A. et al., 2018). Both studies were approved by the Institutional Review Board of Rush University Medical Center, Chicago, IL. As a condition of enrollment in ROS and MAP, participants agreed to annual assessments and brain donation after death. A written informed consent and an anatomical gift act were obtained from each participant. Participants entered the studies without known dementia. This study included autopsied participants with targeted SRM proteomics, neuropathologic data, and R2 images from postmortem MRI that passed quality control. A total of 521 participants with both postmortem MRI and targeted SRM proteomics were used. Among those, 228 (43.8%) were clinically diagnosed with Alzheimer’s dementia, and 355 (68.1%) had pathologic AD. The average age at death was 90.3 years (standard deviation [SD]: 6.1; range 65.9–108.3). A majority were female (70.3%) and almost all were non-Latino whites (95.9%). The average years of education was 15.8 (SD: 3.6; range 3–30).
2.2. Cognitive assessment
A neuropsychological test battery was administered every year with over 95% overall follow-up rate. The cognitive battery consists of 21 tests, 19 in common between MAP and ROS, 17 of which were used to assess multiple domains of cognitive ability, including episodic, semantic, and working memory; perceptual speed; and visuospatial acuity. Each of the scores were z-transformed using baseline means and SDs, and the resulting z-scores from all tests were averaged to derive a composite measure of global cognition. We estimated person-specific rates of cognitive decline based on a linear mixed effects model adjusted for age, sex, and years of education, as previously reported (Yu et al., 2018).
2.3. Neuropathology measures
At autopsy, brains were removed and hemisected in accordance with standard protocol (Schneider et al., 2009). We identified the cerebral hemisphere with more visible pathology mainly based on gross infarcts, and occasionally tumor or hemorrhage. The hemisphere was immersed it in 4% paraformaldehyde solution and refrigerated it at 4° C. The other hemisphere was frozen in preparation for use in various other investigations, including RNA proteomics. Neuropathologies, including AD, cerebral infarcts, Lewy bodies, hippocampal sclerosis, hyperphosphorylated transactive response DNA-binding protein 43 (TDP-43), cerebral amyloid angiopathy (CAA), atherosclerosis, and arteriolosclerosis were evaluated, blind to clinical diagnosis. β-amyloid and PHFtau tangle pathologies were measured using molecular specific immunohistochemistry in eight predetermined brain regions, including superior frontal cortex, dorsolateral prefrontal cortex, entorhinal cortex, and hippocampus, consisting of 20μm sections immunostained with one of three monoclonal antibodies (4G8, 1:9000, Covance Labs, Madison, WI; 6F/3D, 1:50, Dako North America Inc., Carpinteria, CA; and 10D5,1:600, Elan Pharmaceuticals, San Francisco, CA) for β-amyloid, and AT8 (1:1,000, Innogenetics, San Ramon, CA) for PHFtau tangles (Bennett, David A. et al., 2006). β-amyloid load and PHF tau tangle density were obtained by image analysis utilizing Stereo Investigator Version 2017. For β-amyloid, positive regions of interest were manually outlined and analyzed at 20X using an Olympus BX51 microscope attached to a MBF CX9000 camera coupled to a Stage Driver. A grid was placed for sampling about 20–25% area from neocortical regions and 40–45% of the area from entorhinal and hippocampal sector CA1/subiculum cortices. The percent area of β-amyloid immunoreactivity (load) was then estimated using Image J software and averaged across regions. For analyses, amyloid load was defined as the square root of the percent of tissue area that was positive for β-amyloid within each region averaged over all regions for each participant. PHF-tau tangle neurons in each region was quantified after outlining the region of interest and using the optical fractionator probe and random sampling about 10–15% area from neocortex and 15% area from mesial temporal lobe structures. Regional PHF-tau tangle densities were averaged across regions. Macroscopic infarcts were recorded during gross examination and confirmed histologically, and microinfarcts were identified in a minimum of 9 regions per participant using haematoxylin and eosin (H&E) stain (Arvanitakis, Z. et al., 2011). Both chronic macroscopic and microinfarcts were recorded as presence/absence. Lewy bodies in substantial nigra, limbic, and neocortical regions were identified using α-synuclein immunostaining (Schneider et al., 2012) and were recorded as present/absent. Hippocampal sclerosis was assessed using H&E stained sections from mid-hippocampus (Nag et al., 2015) and was recorded as presence/absence. We used monoclonal TDP-43 antibody (Yu, De Jager et al., 2015) to stain 6 regions (amygdala, CA1 of hippocampus, dentate of hippocampus, entorhinal cortex, midtemporal and midfrontal cortices) and manually scanned slides at low power (4X objective) to find the greatest density of TDP-43 inclusions. Objective was then switched to 20 X objective and counts were performed in a 0.25 mm2 sampling frame (500 μm square). All TDP-43 inclusions were included, i.e. neuronal, neuronal intranuclear, and glial cytoplasmic. For these analyses we used the presence or absence of TDP-43 across these 6 regions to stage the severity of progression of TDP-43 from stage 0 (no inclusions), to (1) amygdala only, to (2) extended to other limbic regions to (3) extended to neocortex. Cerebral amyloid angiopathy (CAA) pathology was assessed in 4 neocortical regions: midfrontal, midtemporal, angular, and calcarine cortices. Paraffin-embedded sections were immunostained for β-amyloid using 1 of 3 monoclonal anti-human antibodies: 4G8 (1:9000; Covance Labs, Madison, WI), 6F/3D (1:50; Dako North America Inc., Carpinteria, CA), and 10D5 (1:600; Elan Pharmaceuticals, San Francisco, CA). For CAA assessment, meningeal and parenchymal vessels were assessed for amyloid deposition and scored from 0 to 4, where 0 = no deposition, 1 = scattered segmental but no circumferential deposition, 2 = circumferential deposition up to 10 vessels, 3 = circumferential deposition up to 75% of the region, and 4 = circumferential deposition over 75% of the total region. The CAA score for each region was the maximum of the meningeal and parenchymal CAA scores. Scores were averaged across regions and summarized as a continuous measure of CAA pathology for analyses (Boyle, Patricia A. et al., 2015, Yu, Boyle et al., 2015). Atherosclerosis was assessed by visual inspection of the segmental or circumferential subintimal accumulation of lipid, plasma proteins, and calcium deposition in the circle of Willis, of which the vertebral, basilar, posterior cerebral, middle cerebral, and anterior cerebral arteries and their proximal branches were inspected. Severity of atherosclerosis was graded based on the involvement of each artery and number of arteries from 0 (none) to 6 (severe) (Arvanitakis, Z. et al., Arvanitakis, Zoe et al., 2016). Upon involvement of a new rater, we follow a standard protocol in which the new rater rates 50 to 100 slides that have already been rated by an experienced rater and must be certified on practice slides prior to actual data collection.
The slides are selected to include a representative range of pathology. We then obtain percent of variation in the square-root transformed value of amyloid and tangles due to raters within slide. Generally the variance due to different raters is less than 5%. Similar cross-training is done with TDP-43 data collection, Cerebrovascular, and Lewy body disease pathologies are reviewed by a neuropathologist (Bennett, D. A. et al., 2012).
2.4. Targeted SRM proteomics
Targeted proteomics analysis was performed using frozen tissue from 1,226 dorsolateral prefrontal cortex (DLPFC) samples, the same brain region selected for RNAseq. Details on SRM methods are reported in our previous publications (Andreev et al., 2012, Kim et al., 2014, Petyuk et al., 2010, Yu et al., 2018). Peptide abundance is measured as a ratio of its intensity to the intensity of the spiked-in synthetic stable isotope labeled standard. The endogenous light to labeled heavy peptide ratios were log2-transformed and median-centered. This effectively made the log2 abundance ratios relative to the median abundance. The reproducibility and the quality of peptide quantification were assessed using repetitive measurements of pooled samples employed as technical controls. We considered a peptide signal informative if the variance, both technical and biological, across the individual brain samples exceeded 2-fold of technical variance. For proteins with multiple peptides quantified, the peptide with the highest signal to noise ratio (total/technical variance) was used.
2.5. MRI Acquisition
At approximately one month postmortem (mean=30 days; SD=19.2 days), the refrigerated hemisphere at 4 °C from each decedent was imaged using a 3-Tesla MRI scanner after allowing the tissue to warm to room temperature. Four different scanners were used to acquire fast spin-echo T2-weighted MRI data with at least two different echo times (TE). Details of MRI acquisition have been previously reported (Dawe et al., 2009, Dawe et al., 2014a, Dawe et al., 2016, Dawe et al., 2018). The transverse relaxation time constant, T2, describing the decay rate of the transverse component of magnetization, is sensitive to, among other things, brain tissue’s free water content and the presence of paramagnetic materials, such as iron. As previously described (Dawe et al., 2014a, Dawe et al., 2016, Dawe et al., 2018), we quantified the reciprocal of T2, R2, to reduce skewness and achieve a more normally distributed signal. R2 maps were spatially registered to a cerebral hemisphere template, using first linear and then nonlinear transformations (Dawe et al., 2016). In order to account for inter-scanner differences in R2, we carried out a voxel-by-voxel normalization of R2 values within each group of hemispheres imaged on a given scanner by subtracting the median and dividing by the interquartile range of R2 for that scanner. In addition, we carried out spatial smoothing with full width at half maximum 2mm. Details of postmortem MRI preprocessing procedure have been described previously (Dawe et al., 2014a, Dawe et al., 2016, Dawe et al., 2018).
2.6. Regions in the postmortem brain associated with individual proteins
Voxel-wise linear regression analysis was applied to each half-brain (number of voxels ≥ 4×105) to identify brain regions where postmortem R2 was associated with abundance of each of our four proteins - AK4, ITPK1, IGFBP5, HSPB2 - after controlling for age at death, sex, and years of education. Thresholds were sequentially applied, first on individual voxels (p<0.01) and then on individual clusters (p <0.05 corrected for multiple comparisons) based on Random Field (RF) theory (Friston et al., 1994, Worsley et al., 1996), a part of Statistical Parametric Mapping (SPM). Principal component analysis (PCA), based on correlations of mean R2 values between clusters, was used to reduce dimensionality (or number of clusters). For an instance, the first PC is sufficient to explain total variation in R2 clusters if all the clusters are highly mutually correlated. The number of PC’s chosen was based on the proportion of variance explained. Assignment of individual clusters into groups was based on loadings of PC’s. We illustrate this procedure with AK4 from Supplementary material 1. The first two PCs explained 93% of the variance while the 3rd PC only explained 7%. Therefore, the first two PCs were included in analyses. In PC1, Cluster 1 and 2 had almost equal loadings while the loading of Cluster 3 was one tenth of those. Cluster 1 and 2 were therefore assigned to PC1. The loading of Cluster 3 suggested that it should be included in PC2. Such disjoint sets are referred to as cluster groups. The mean value of postmortem R2 from each cluster group was utilized for subsequent mediation analysis. It is noteworthy that our mediation analysis is a simplified approach with clusters while voxel-wise mediation approach is also available with intensive computation.
2.7. Mediation analysis
We examined three models: (a) cognitive decline as a function of three demographic variables (age at death, sex, and years of education) and protein abundance; (b) postmortem R2 as a function of the three demographic variables and protein abundance, and (c) cognitive decline as a function of the three demographic variables, protein abundance, and postmortem R2. Since postmortem R2 is known to be variably correlated with neuropathology, we repeated the analysis and included neuropathology measures in each step. In the mediation analysis, we tested the effect of each protein on cognitive decline through the aforementioned groups of R2 clusters, also known as the indirect effect. The remaining effect of each protein on cognitive decline not acting through R2 after controlling covariates - the direct effect - was calculated and the sum of these two effects constituted the total effect of each protein on cognitive decline, after controlling covariates. In the presence of multiple groups of R2 clusters as mediators, we assessed individual indirect effects by individual mediators. Mediation analysis was performed by SAS macro provided by Hayes & Preacher (Hayes and Preacher, 2014).
3. Results
3.1. AK4, HSPB2, and IGHBP5 with a faster rate of cognitive decline
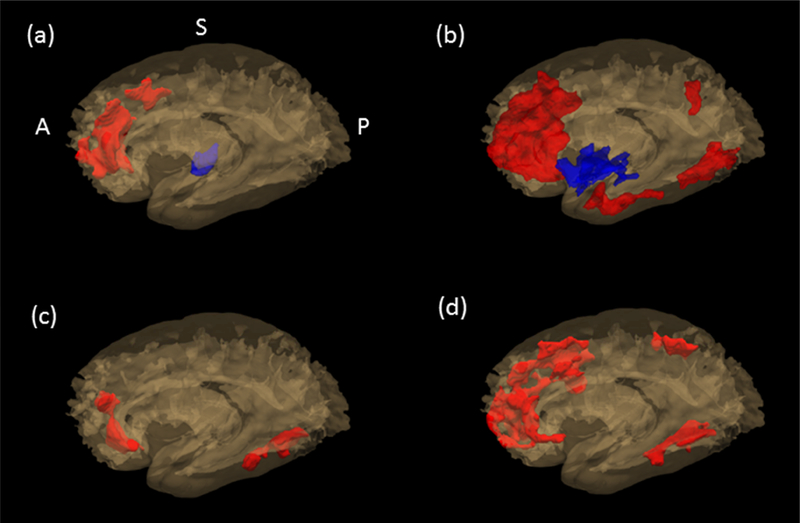
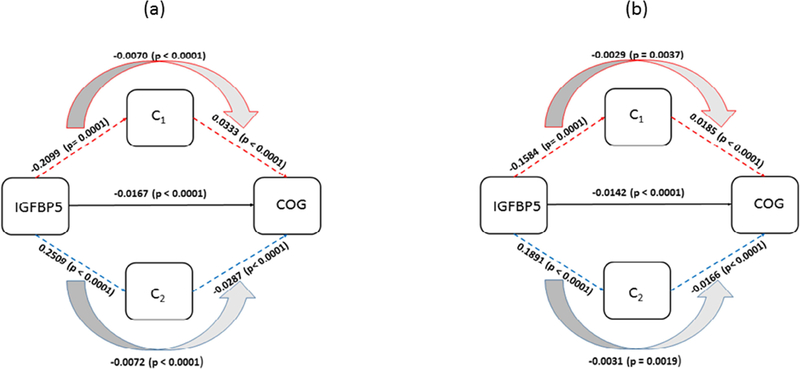
Voxel-wise postmortem R2 analysis identified areas in the brain associated with abundance of each protein. Three clusters with a total volume of 6563 (mm3) in the postmortem brain template were identified for AK4, six clusters with a total volume of 29535 (mm3) for IGFBP5, and two clusters with a total volume of 4464 (mm3) for HSPB2. As shown in Figure 1, clusters were predominantly located in frontal, temporal, and parietal areas and exhibited a high degree of overlap. Based on the PCA results, the first PC for HSPB2, explaining 75.2% of the total variance, was chosen; For AK4 and IGFBP5, the first two PCs, explaining 92.9% and 73.0% of the total variance, respectively, were chosen. It is noteworthy that clusters grouped together were consistent in their major tissue types (white versus grey matter). Of the two cluster groups respectively for AK4 and IGFBP5, the cluster group 1 were mainly located in the white matter, and the cluster group 2 in the grey matter while all the clusters from HSPB2 were in the white matter. By tissue types, lower R2 cluster in white matter and higher R2 cluster in grey matter was associated with more rapid cognitive decline. Individual groups of clusters are shown in Figure 2 (a) through (c). In each panel, red-colored areas are for clusters mainly located in white matter, and blue-colored areas are for clusters in grey matter.
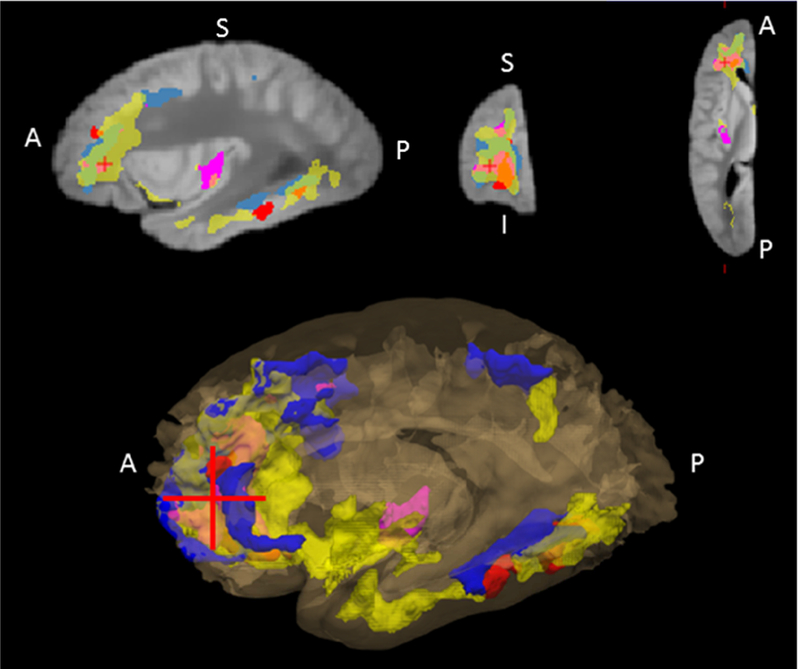
Figure 1.
Postmortem brain areas associated with each of the four proteins, AK4, ITPK1, IGFBP5, and HSPB2. A 3-D view of all the clusters from the four proteins is shown in bottom panel, and its sagittal, coronal, and axial view at the red crosshair is shown in the top panel. The regions associated with AK4 is colored in pink, ITPK1 in blue, IGFBP5 in yellow, and HSPB2 in red.
Figure 2.
Map of individual group of clusters associated with each of the four proteins, AK4, ITPK1, IGFBP5, and HSPB2. AK4 is shown in (a), IGFBP5 in (b), HSPB2 in (c), and ITPK1 in (d). Clusters grouped together were consistent in their major tissue types (white vs. grey). In each figure, red-colored area shows the cluster group located in white matter, and the blue-colored area in grey matter.
In mediation analysis, we examined the mediating role of the postmortem R2 clusters on the relationship between each of the proteins and cognitive decline, adjusting for age, sex, and education. Higher AK4, HSPB2, and IGFBP5 were associated with faster decline in cognition (AK4: β= −0.0173, p<0.0001; HSPB2: β= −0.0138, p=0.0013; IGFBP5: β= −0.0309, p<0.0001) (Table 1a). The association between each protein and R2 clusters is shown in Table 1b. All three proteins showed significant indirect effects on cognitive decline through postmortem R2 clusters (Table 1c). The proportion of indirect effects out of the total effect for each protein was about 50% (AK4– 47%; IGFBP5– 45 %; HSPB2– 50%; Table 3).
Table 1a.
Association of Protein on cognitive decline after controlling demographic variables
| Beta (total effect) | SE | P | |
|---|---|---|---|
| AK4 | −0.0173 | 0.0043 | <0.0001 |
| ITPK1 | 0.0147 | 0.0043 | 0.0006 |
| IGFBP5 | −0.0309 | 0.0041 | <0.0001 |
| HSPB2 | −0.0138 | 0.0043 | 0.0013 |
Model: Cognitive decline = Demographic variables + Protein
Table 1b.
Association of Protein on postmortem ex vivo R2 after controlling demographic variables
| Cluster group 1 | Cluster group 2 | |||||
|---|---|---|---|---|---|---|
| Beta | SE | P | Beta | SE | P | |
| AK4 | −0.1520 | 0.0431 | 0.0004 | 0.170 | 0.0436 | 0.0001 |
| ITPK1 | 0.2002 | 0.0425 | <0.0001 | - | - | - |
| IGFBP5 | −0.2099 | 0.0423 | <0.0001 | 0.2509 | 0.0425 | <0.0001 |
| HSPB2 | −0.1876 | 0.0421 | <0.0001 | |||
Model: Postmortem R2 = Demographic variables + Protein
Note: Cluster group 1 was mainly located in white matter while cluster group 2 was in grey matter.
Table 1c.
Association of protein on cognitive decline after controlling demographic variables and postmortem R2
| Protein | Cluster group1 | Cluster group2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | se | P | Beta | se | P | Beta | se | P | |||
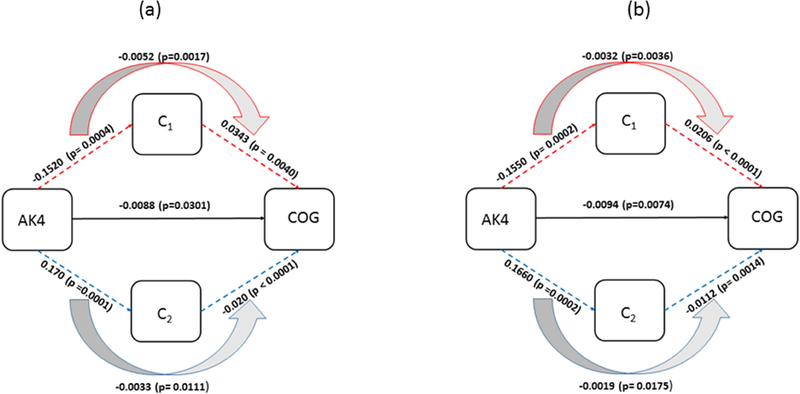
| AK4 | Direct effect | −0.0088 | 0.0040 | 0.0301 | 0.0343 | 0.0040 | <0.0001 | −0.020 | 0.0040 | <0.0001 | |
| Indirect effect | through C1 | −0.0052 | 0.0017 | 0.0022 | |||||||
| Indirect effect | through C2 | −0.0033 | 0.0013 | 0.0111 | |||||||
| ITPK1 | Direct effect | 0.0084 | 0.0041 | 0.0420 | 0.0315 | 0.0042 | <0.0001 | - | - | - | |
| Indirect Effect | through C1 | 0.0063 | 0.0015 | <0.0001 | |||||||
| IGFBP5 | Direct effect | −0.0167 | 0.0039 | <0.0001 | 0.0333 | 0.0039 | <0.0001 | −0.0287 | 0.0039 | <0.0001 | |
| Indirect Effect | through C1 | −0.0070 | 0.0016 | <0.0001 | |||||||
| Indirect Effect | through C2 | −0.0072 | 0.0015 | <0.0001 | |||||||
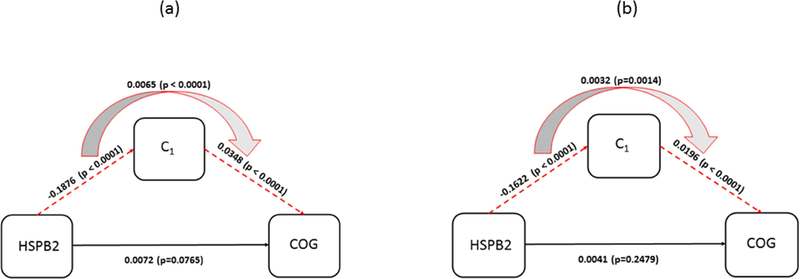
| HSPB2 | Direct effect | −0.0072 | 0.0041 | 0.0765 | 0.0348 | 0.0042 | <0.0001 | - | - | - | |
| Indirect Effect | through C1 | −0.0065 | 0.0016 | <0.0001 | |||||||
Model: Cognitive decline = Demographic variables + Postmortem R2 + Protein
Note: Cluster group 1 was mainly located in white matter while cluster group 2 was in grey matter.
Table 3.
Proportion of the direct and the indirect effect to the total effect on cognition by protein
| Covariates | Controlling only demographic variables | Controlling both of demographic and pathology variables | |||
|---|---|---|---|---|---|
| Effect | Effect | proportion | Effect | proportion | |
| AK4 | total | −0.017 | 100.0% | −0.015 | 100.0% |
| direct | −0.009 | 52.9% | −0.009 | 60.0% | |
| indirect through R2 | −0.008 | 47.1% | −0.006 | 40.0% | |
| ITPK1 | total | 0.015 | 100.0% | 0.010 | 100.0% |
| direct | 0.008 | 53.3% | 0.007 | 70.0% | |
| indirect through R2 | 0.007 | 46.7% | 0.003 | 30.0% | |
| IGFBP5 | total | −0.031 | 100.0% | −0.020 | 100.0% |
| direct | −0.017 | 54.8% | −0.014 | 70.0% | |
| indirect through R2 | −0.014 | 45.2% | −0.006 | 30.0% | |
| HSPB2 | total | −0.014 | 100.0% | −0.007 | 100.0% |
| direct | −0.007 | 50.0% | −0.004 | 57.1% | |
| indirect through R2 | −0.007 | 50.0% | −0.003 | 42.9% | |
Since microstructural changes in the brain measured by postmortem R2 is known to be correlated with various neuropathology measurements (Dawe et al., 2014a, Dawe et al., 2016, Dawe et al., 2018, House et al., 2008), we next examined the mediating role of microstructural changes in the brain measured by postmortem R2 in the relationship between each of the three proteins (AK4, IGFBP5, and HSPB2) and cognitive decline, after adjusting for AD and other common neuropathologies, as well as age, sex, and education (Table 2a through 2c). Similar to the results of analyses not including neuropathology measures, the mediation effect of microstructural changes in the brain measured by postmortem R2 on association between protein abundance and cognitive decline remained significant (Table 2c). The proportion of indirect effects to the total effect for each protein was about 40% (AK4– 40%; IGFBP5– 30%; HSPB2– 43%; Table 3). Diagrams for each mediation analysis is shown in Figure 3 through 5.
Table 2a.
Association of protein on cognitive decline after controlling demographic variables and neuropathology
| Beta (total effect) | SE | P | |
|---|---|---|---|
| AK4 | −0.0145 | 0.0035 | <0.0001 |
| ITPK1 | 0.0101 | 0.0035 | 0.0050 |
| IGFBP5 | −0.0203 | 0.0035 | <0.0001 |
| HSPB2 | −0.0073 | 0.0036 | 0.0418 |
Model: Cognitive decline = Demographic variables + Neuropathology + Protein
Table 2c.
Association of protein on cognitive decline after controlling demographic variables, neuropathology, and postmortem R2
| Protein | Cluster group1 | Cluster group2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | se | P | Beta | se | P | Beta | se | P | |||
| AK4 | Direct effect | −0.0094 | 0.0035 | 0.0074 | 0.0206 | 0.0037 | <0.0001 | −0.0112 | 0.0035 | 0.0014 | |
| Indirect Effect | through C1 | −0.0032 | 0.0011 | 0.0036 | |||||||
| Indirect Effect | through C2 | −0.0019 | 0.0008 | 0.0175 | |||||||
| ITPK1 | Direct effect | 0.0075 | 0.0036 | 0.0362 | 0.0147 | 0.0038 | 0.0001 | - | - | - | |
| Indirect Effect | through C1 | 0.0025 | 0.0009 | 0.0077 | |||||||
| IGFBP5 | Direct effect | −0.0142 | 0.0035 | <0.0001 | 0.0185 | 0.0038 | <0.0001 | −0.0166 | 0.0036 | <0.0001 | |
| Indirect Effect | through C1 | −0.0029 | 0.0010 | 0.0037 | |||||||
| Indirect Effect | through C2 | −0.0031 | 0.0010 | 0.0019 | |||||||
| HSPB2 | Direct effect | −0.0041 | 0.0035 | 0.2479 | 0.0196 | 0.0038 | <0.0001 | - | - | ||
| Indirect Effect | through C1 | −0.0032 | 0.0010 | 0.0014 | |||||||
Model: Cognitive decline = Demographic variables + Neuropathology + Postmortem R2 + Protein
Note: Cluster group 1 was mainly located in white matter while cluster group 2 was in grey matter.
Figure 3.
Diagram for mediation of postmortem R2 clusters (C1, C2) on association between AK4 and cognitive decline. (a) The effects estimated with demographic variables as covariates are shown. (b) The effects estimated with AD and common neuropathology added to the covariates in (a) are shown. C1 and C2 are mean of the R2 measurements from cluster groups that are associated with AK4.
Figure 5.
Diagram for mediation of postmortem R2 on association between HSPB2 and cognitive decline. (a) The effects estimated with demographic variables as covariates are shown (b) The effects with AD and common neuropathology added to the covariates in (a) are shown. C1 is mean of R2 measurements from the cluster group that is associated with HSPB2.
3.2. ITPK1 with a slower rate of cognitive decline
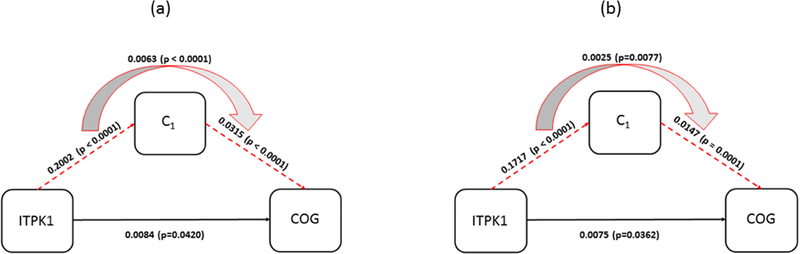
Voxel-wise postmortem R2 analysis identified areas in the brain associated with ITPK1 abundance in three clusters, involving a total volume of 13611 (mm3). These clusters were all involved in the first PC with almost equal weights (accounting for 74.5% of the total variance), and mainly located in white matter. Mediation effects of microstructural changes in the brain measured by postmortem R2 on the relationship between ITPK1 and cognitive decline, adjusting for age, sex, and education, was tested. Higher ITPK1 abundance was associated with slower decline (β= 0.0147, p=0.0006; Table 1a). ITPK1 showed a significant indirect effect on cognitive decline through postmortem R2 clusters (β= 0.0084, p= 0.042; Table 1c). Proportion of indirect effect to the total effect of ITPK1was 47% (Table 3). The mediation effect of postmortem R2 clusters on the relationship between ITPK1 and cognitive decline, adjusting for AD and other common neuropathologies, in addition to age, sex, and education, is shown in Table 2a through 2c. Similar to our previous results, the mediation effect of postmortem R2 clusters remained significant (Table 2c). Diagram for each mediation analysis is shown in Figure 6.
Figure 6.
Diagram for mediation of postmortem R2 on association between ITPK1 and cognitive decline. (a) The effects estimated with demographic variables as covariates are shown. (b) The effects with AD and common neuropathology added to the covariates in (a) are shown. C1 is mean of R2 measurements from the cluster group that is associated with ITPK1.
4. Discussion
We have previously reported that higher expression of the coexpressed gene set m109 results in greater ß-amyloid pathology burden and more rapid cognitive decline, and these reports proposed two genes, INPPL1 and PLXNB1, as potential drivers of the network (Mostafavi et al., 2018). In a subsequent study, PLXNB1’s involvement was further validated in 834 human autopsy samples showing higher levels of PLXNB1 contributed to increased accumulation of ß-amyloid and PHFtau tangle density (Yu et al., 2018). M109, however, also has a large direct edge to cognitive decline. We found four proteins associated with cognitive decline that were not explained by AD and other pathologies: AK4, ITPK1, HSPB2, and IGFBP5 (Yu et al., 2018). In this study, we demonstrate that changes in the brain measured by postmortem R2 mediates the relationship between each protein and cognitive decline. Specifically, higher abundances of AK4, HSPB2, and IGFBP5 are associated with faster cognitive decline, as mediated by postmortem R2 clusters in extensive white matter regions; lower R2 is a marker of neurodegenerative/vascular disease within white matter (Dickie et al., 2016, Fleischman et al., 2015, Gebeily et al., 2014, Williamson et al., 2018). Conversely, higher abundance of ITPK1 was associated with slower cognitive decline as mediated by postmortem R2 clusters in white matter, where higher white matter R2 is a marker of tissue health. In a previous study (Yu et al., 2018), AK4 and ITPK1 abundances present a degree of resilience effect that is defined as changes in cognitive function not explained by known pathologies (Boyle, P. A. et al., 2013). Out of these two proteins, we found that ITPK1 is of particular interest, in that its higher abundance inhibits cognitive decline through increased white matter fidelity. This protective effect could be related to other factors, such as regular participation in cognitively stimulating activities, which has been shown by diffusion MRI to promote white matter microstructural integrity (Arfanakis, K. et al., 2016). Thus, further studies on the modulating effect of ITPK1 on various risk/resilience factors in cognitive aging are warranted. If these proteins play a causal role in regulating brain structure, it may be possible to define their mechanisms of action through existing molecular studies. AK4 is a mitochondrial protein from the adenylate kinase family, whose members are involved in ubiquitous energy metabolism and homeostasis of cellular adenine nucleotide composition (Yoneda et al., 1998). A prior study found high levels of endogenous AK4 in hypoxia-treated cells and spinal cords of an ALS mouse model, suggesting enzymatically inactive AK4 is a stress responsive protein critical to cell survival and proliferation (Liu et al., 2009). HSPB2 belongs to a family of heat shock proteins (HSPs), originally discovered as stress-inducible proteins, and newly emerged as a regulator of physiological functions like protein management and cell death (Stetler et al., 2010) in the central nervous system. IGFBP5 is a binding protein that regulates activity of insulin-like growth factors (IGF), which plays a crucial role in neurodevelopment and apoptosis. Evidence from mouse models show that neuronal IGFBP5 overexpression results in motor neuron degeneration and myelination defects (Marshman et al., 2003, O’Kusky and Ye, 2012, Simon et al., 2015). ITPK1 is a regulatory enzyme that plays a key role in intracellular inositol phosphate (IP) metabolism, a process that plays crucial roles in diverse cellular functions like growth, apoptosis, migration, and differentiation. Overexpression of ITPK1 leads to increased levels of IP4, and highly phosphorylated forms of inositol in mammalian cells, including IP5, IP6. In mice, reduced levels of ITPK1 result in the development of neural tube defects (Majerus et al., 2010, Wilson et al., 2009).
Viewing these results in light of existing literature on the function of individual proteins offers plausible mechanisms underlying the relationships among protein abundance, brain microstructure, and cognitive decline. This evidence is consistent with the directionality of our findings, namely that abundant AK4, HSPB2, and IGFBP5 are linked with faster cognitive decline via decreased brain microstructural integrity, as marked by lower R2. In addition, hypoxia-driven demyelination is a plausible factor underlying R2 depression (Dawe et al., 2016). Association of ITPK1 with cognitive decline is however different from AK4, IGFBP5, and HSPB2. Namely, abundant ITPK1 seems to support rather than detract from brain tissue integrity, which would likely help to preserve cognitive abilities while manifesting as higher R2 in extensive frontal, temporal, and parietal white matter areas. This is consistent with the fact that ITPK1 is associated with a slower rate of cognitive decline.
We hypothesized the mediation model tested in this study based on our previous study (Mostafavi et al., 2018), in which we showed that m109, a network of gene transcripts, mathematically has a causal relation with β-amyloid, and a separate effect on cognitive decline independent of β-amyloid and PHFtau pathology. It was also found that manipulating some m109 genes in human astrocyte cell cultures of β-amyloid production recapitulates the predictions from the model supporting causality. The proteins of interest in this study were nominated from m109 and found to be associated with cognitive decline controlling for common brain pathologies (Yu et al., 2018). Here, we replace common brain pathologies with R2, a proxy biomarker of microstructural changes in the brain some of which are beneficial and others deleterious. Our findings are consistent with but do not prove a causal relationship.
We examined the association of APOE genotype – a well-known risk factor for cognitive decline as well as Alzheimer’s dementia – with the expression level of the four proteins. The mediation models were subsequently augmented with terms for APOE ε4 genotype and APOE protein and the results were not materially changed (data not shown).
R2 – the inverse of the transverse relaxation time constant, T2 – describes the decay rate of the transverse component of magnetization. R2 in vivo imaging is influenced by a number of factors, including the ratio of free to myelin-bound water molecules, and the presence of paramagnetic molecules, such as the iron-laden compounds ferritin and hemosiderin (Dawe et al., 2014b). Unlike in vivo imaging, natural tissue decomposition and chemical fixation cause the postmortem tissue’s MRI properties to change as postmortem fixation time elapses (Dawe et al., 2009). Although we timed the vast majority of our postmortem scans to occur at a time postmortem when R2 values throughout the brain should be relatively stable (approximately 30 days postmortem), we also conducted sensitivity analysis with postmortem fixation time elapsed as a covariate. The median postmortem fixation time elapsed was 33.7 days (mean 39.6, SD 15.1, min 22.2, max 125.9). Each step of mediation analysis was repeated with postmortem fixation time elapsed. Results were highly consistent with the results shown in Table 1 and 2.
Abundance of the four proteins from m109 in the brain were explained in part by microstructural changes in the brain measured by postmortem R2, which accounts for a significant amount of the variance in cognitive decline. Thus, it provides a molecular anchor to an important neuroimaging feature of the aging brain. In addition, regions of the brain associated with abundances of each of the proteins overlapped extensively, which is consistent with these proteins being functionally related to one another; they were originally identified to be coexpressed at the RNA level in the same cortex, DLPFC (Mostafavi et al., 2018). We also found that the four proteins showed association with R2 in areas beyond DLPFC in the postmortem brain. We previously reported R2 associations with areas in the postmortem brain beyond DLPFC with transcriptomic and epigenomic data as well (Gaiteri et al., 2018, Yu et al., 2017). This is expected since DLPFC is an important hub in many functional brain networks. It is likely that such associations are best detected in normal appearing tissue in MRI but not visible lesions. Unfortunately, spatial variation in abundance of each protein across postmortem brain is unknown and warrants further investigation.
The study has several strengths. The community-based cohort enrolls persons without dementia. The high clinical follow-up and autopsy rates result in high internal validity. Our study is based on cohorts that have relatively large sample size of longitudinal cognitive function data, and the wide range of neuropathologies. However, the study also has several limitations. We select the hemisphere less affected by visible pathology for proteomics. We acknowledge that such a nonrandom selection may cause a bias in findings most likely towards the null. We also acknowledge the limitations of drawing causal inferences from cross-sectional associations. We note that, to strictly assess causal inference, it warrants further studies by manipulating those proteins in an ex vivo model system of brain microstructure. We utilized only postmortem R2 images of which mechanism can only be attributed to increased water or iron accumulation. Further research is warranted to link these proteins to measures from ante-mortem imaging modalities, such as diffusion tensor imaging and resting state functional MRI, to explain structural connection between areas in the brain and the functional connectivity.
Supplementary Material
Figure 4.
Diagram for mediation of postmortem R2 clusters (C1, C2) on association between IGFBP5 and cognitive decline. (a) The effects estimated with demographic variables as covariates are shown. (b) The effects estimated with AD and common neuropathology added to the covariates in (a) are shown. C1 and C2 are mean of R2 measurements from each cluster group that is associated with IGFBP5.
Table 2b.
Association of protein on postmortem ex vivo R2 after controlling demographic variables and neuropathology
| Cluster group 1 | Cluster group 2 | |||||
|---|---|---|---|---|---|---|
| Beta | SE | P | Beta | SE | P | |
| AK4 | −0.1550 | 0.0407 | 0.0002 | 0.1660 | 0.0438 | 0.0002 |
| ITPK1 | 0.1717 | 0.0406 | <0.0001 | - | - | - |
| IGFBP5 | −0.1584 | 0.0414 | 0.0001 | 0.1891 | 0.0427 | <0.0001 |
| HSPB2 | −0.1622 | 0.0402 | <0.0001 | - | - | - |
Model: Postmortem R2 = Demographic variables + Neuropathology + Protein
Note: Cluster group 1 was mainly located in white matter while cluster group 2 was in grey matter.
ACKNOWLEDGEMENT
We give thanks to all the ROS and MAP participants, and also all the staffs and investigators at the Rush Alzheimer’s Disease Center (RADC) for providing and processing high quality data. In addition, we give thanks to Dr. Katherine Blizinsky for helpful discussions and comments. Please visit the RADC Research Resource Sharing Hub (www.radc.rush.edu) to obtain data for research purpose. This study was supported in part by NIH grants P30R01AG10161, R01AG15819, R01AG17917, U01AG046152, R01AG034374, 1R01AG057911, U01AG61356.
Footnotes
POTENTIAL CONFLICTS OF INTEREST
Nothing to report
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreev VP, Petyuk VA, Brewer HM, Karpievitch YV, Xie F, Clarke J, Camp D, Smith RD, Lieberman AP, Albin RL, Nawaz Z, El Hokayem J, Myers AJ, 2012. Label-free quantitative LC-MS proteomics of Alzheimer’s disease and normally aged human brains. J.Proteome Res 11, 3053–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfanakis K, Wilson RS, Barth CM, Capuano AW, Vasireddi A, Zhang S, Fleischman DA, Bennett DA, 2016. Cognitive activity, cognitive function, and brain diffusion characteristics in old age. Brain Imaging Behav. 10, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arfanakis K, Gui M, Tamhane AA, Carew JD, 2007. Investigating the Medial Temporal Lobe in Alzheimer’s Disease and Mild Cognitive Impairment, with Turboprop Diffusion Tensor Imaging, MRI-volumetry, and T2-relaxometry. 1, 11–21. [Google Scholar]
- Arvanitakis Z, Capuano AW, Leurgans SE, Buchman AS, Bennett DA, Schneider JA,. The Relationship of Cerebral Vessel Pathology to Brain Microinfarcts. [DOI] [PMC free article] [PubMed]
- Arvanitakis Z, Leurgans SE, Barnes LL, Bennett DA, Schneider JA, 2011. Microinfarct pathology, dementia, and cognitive systems. Stroke. 42, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitakis Z, Capuano AW, Leurgans SE, Bennett DA, Schneider JA, 2016. Relation of cerebral vessel disease to Alzheimer’s disease dementia and cognitive function in elderly people: a cross-sectional study. 15, 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Buchman AS, Boyle PA, Barnes LL, Wilson RS, Schneider JA, 2018. Religious Orders Study and Rush Memory and Aging Project. J.Alzheimers Dis. 64, S161–S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Wilson RS, Boyle PA, Buchman AS, Schneider JA, 2012. Relation of neuropathology to cognition in persons without cognitive impairment. Ann.Neurol. 72, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Schneider JA, Tang Y, Arnold SE, Wilson RS, 2006. The effect of social networks on the relation between Alzheimer’s disease pathology and level of cognitive function in old people: a longitudinal cohort study. 5, 406–412. [DOI] [PubMed] [Google Scholar]
- Boyle PA, Wilson RS, Yu L, Barr AM, Honer WG, Schneider JA, Bennett DA, 2013. Much of late life cognitive decline is not due to common neurodegenerative pathologies. Ann.Neurol. 74, 478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle PA, Yu L, Nag S, Leurgans S, Wilson RS, Bennett DA, Schneider JA, 2015. Cerebral amyloid angiopathy and cognitive outcomes in community-based older persons. Neurology. 85, 1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RJ, Bennett DA, Schneider JA, Leurgans SE, Kotrotsou A, Boyle PA, Arfanakis K, 2014a. Ex vivo T2 relaxation: associations with age-related neuropathology and cognition. Neurobiol.Aging. 35, 1549–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RJ, Bennett DA, Schneider JA, Leurgans SE, Kotrotsou A, Boyle PA, Arfanakis K, 2014b. Ex vivo T2 relaxation: associations with age-related neuropathology and cognition. Neurobiol.Aging. 35, 1549–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RJ, Bennett DA, Schneider JA, Vasireddi SK, Arfanakis K, 2009. Postmortem MRI of human brain hemispheres: T2 relaxation times during formaldehyde fixation. Magn.Reson.Med 61, 810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RJ, Yu L, Leurgans SE, Schneider JA, Buchman AS, Arfanakis K, Bennett DA, Boyle PA, 2016. Postmortem MRI: a novel window into the neurobiology of late life cognitive decline. Neurobiol.Aging 45, 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RJ, Yu L, Schneider JA, Arfanakis K, Bennett DA, Boyle PA, 2018. Postmortem brain MRI is related to cognitive decline, independent of cerebral vessel disease in older adults. Neurobiol.Aging. 69, 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie DA, Ritchie SJ, Cox SR, Sakka E, Royle NA, Aribisala BS, Valdés Hernández MDC, Maniega SM, Pattie A, Corley J, Starr JM, Bastin ME, Deary IJ, Wardlaw JM, 2016. Vascular risk factors and progression of white matter hyperintensities in the Lothian Birth Cohort 1936. Neurobiol.Aging. 42, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischman DA, Yang J, Arfanakis K, Arvanitakis Z, Leurgans SE, Turner AD, Barnes LL, Bennett DA, Buchman AS, 2015. Physical activity, motor function, and white matter hyperintensity burden in healthy older adults. Neurology. 84, 1294–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Worsley KJ, Frackowiak RS, Mazziotta JC, Evans AC, 1994. Assessing the significance of focal activations using their spatial extent. Hum.Brain Mapp. 1, 210–220. [DOI] [PubMed] [Google Scholar]
- Fulcher BD, Fornito A, 2016. A transcriptional signature of hub connectivity in the mouse connectome. Proc.Natl.Acad.Sci.USA. 113, 1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Dawe R, Mostafavi S, Blizinsky KD, Tasaki S, Komashko V, Yu L, Wang Y, Schneider JA, Arfanakis K, De Jager PL, Bennett DA, 2018. Gene expression and DNA methylation are extensively coordinated with MRI-based brain microstructural characteristics. Brain Imaging Behav. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebeily S, Fares Y, Kordahi M, Khodeir P, Labaki G, Fazekas F, 2014. Cerebral white matter hyperintensities (WMH): an analysis of cerebrovascular risk factors in Lebanon. Int.J.Neurosci 124, 799–805. [DOI] [PubMed] [Google Scholar]
- Hayes AF, Preacher KJ, 2014. Statistical mediation analysis with a multicategorical independent variable. Br.J.Math.Stat.Psychol 67, 451–470. [DOI] [PubMed] [Google Scholar]
- House MJ, St Pierre TG, Foster JK, Martins RN, Clarnette R, 2006. Quantitative MR imaging R2 relaxometry in elderly participants reporting memory loss. AJNR Am.J.Neuroradiol 27, 430–439. [PMC free article] [PubMed] [Google Scholar]
- House MJ, St Pierre TG, McLean C, 2008. 1.4T study of proton magnetic relaxation rates, iron concentrations, and plaque burden in Alzheimer’s disease and control postmortem brain tissue. Magn.Reson.Med 60, 41–52. [DOI] [PubMed] [Google Scholar]
- Kim M, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, Thomas JK, Muthusamy B, Leal-Rojas P, Kumar P, Sahasrabuddhe NA, Balakrishnan L, Advani J, George B, Renuse S, Selvan LDN, Patil AH, Nanjappa V, Radhakrishnan A, Prasad S, Subbannayya T, Raju R, Kumar M, Sreenivasamurthy SK, Marimuthu A, Sathe GJ, Chavan S, Datta KK, Subbannayya Y, Sahu A, Yelamanchi SD, Jayaram S, Rajagopalan P, Sharma J, Murthy KR, Syed N, Goel R, Khan AA, Ahmad S, Dey G, Mudgal K, Chatterjee A, Huang T, Zhong J, Wu X, Shaw PG, Freed D, Zahari MS, Mukherjee KK, Shankar S, Mahadevan A, Lam H, Mitchell CJ, Shankar SK, Satishchandra P, Schroeder JT, Sirdeshmukh R, Maitra A, Leach SD, Drake CG, Halushka MK, Prasad TSK, Hruban RH, Kerr CL, Bader GD, Iacobuzio-Donahue C, Gowda H, Pandey A, 2014. A draft map of the human proteome. Nature. 509, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch SJ, Jacobs RW, Butcher LL, Beatty J, 1992. Prolongation of magnetic resonance T2 time in hippocampus of human patients marks the presence and severity of Alzheimer’s disease. Neurosci.Lett 134, 187–190. [DOI] [PubMed] [Google Scholar]
- Krienen FM, Yeo BT, Ge T, Buckner RL, Sherwood CC, 2016. Transcriptional profiles of supragranular-enriched genes associate with corticocortical network architecture in the human brain. Proc.Natl.Acad.Sci.U.S.A 113, E469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Strom AL, Zhai J, Gal J, Bao S, Gong W, Zhu H, 2009. Enzymatically inactive adenylate kinase 4 interacts with mitochondrial ADP/ATP translocase. Int.J.Biochem.Cell Biol 41, 1371–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW, Wilson DB, Zhang C, Nicholas PJ, Wilson MP, 2010. Expression of inositol 1,3,4-trisphosphate 5/6-kinase (ITPK1) and its role in neural tube defects. Adv.Enzyme Regul 50, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshman E, Green KA, Flint DJ, White A, Streuli CH, Westwood M, 2003. Insulin-like growth factor binding protein 5 and apoptosis in mammary epithelial cells. J.Cell.Sci 116, 675. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Gaiteri C, Sullivan SE, White CC, Tasaki S, Xu J, Taga M, Klein HU, Patrick E, Komashko V, McCabe C, Smith R, Bradshaw EM, Root DE, Regev A, Yu L, Chibnik LB, Schneider JA, Young-Pearse TL, Bennett DA, De Jager PL, 2018. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat.Neurosci 21, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, Yu L, Capuano AW, Wilson RS, Leurgans SE, Bennett DA, Schneider JA, 2015. Hippocampal sclerosis and TDP-43 pathology in aging and Alzheimer disease. Ann.Neurol 77, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kusky J, Ye P, 2012. Neurodevelopmental effects of insulin-like growth factor signaling. Front.Neuroendocrinol 33, 230–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petyuk VA, Qian WJ, Smith RD, Smith DJ, 2010. Mapping protein abundance patterns in the brain using voxelation combined with liquid chromatography and mass spectrometry. Methods. 50, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T, Barker GJ, Bokde AL, Bromberg U, Buchel C, Conrod P, Fauth-Buhler M, Flor H, Frouin V, Gallinat J, Garavan H, Gowland P, Heinz A, Lemaitre H, Mann KF, Martinot JL, Nees F, Paus T, Pausova Z, Rietschel M, Robbins TW, Smolka MN, Spanagel R, Strohle A, Schumann G, Hawrylycz M, Poline JB, Greicius MD, IMAGEN consortium, 2015. BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science. 348, 1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JA, Arvanitakis Z, Leurgans SE, Bennett DA, 2009. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann.Neurol. 66, 200–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JA, Arvanitakis Z, Yu L, Boyle PA, Leurgans SE, Bennett DA, 2012. Cognitive impairment, decline and fluctuations in older community-dwelling subjects with Lewy bodies. Brain. 135, 3005–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon CM, Rauskolb S, Gunnersen JM, Holtmann B, Drepper C, Dombert B, Braga M, Wiese S, Jablonka S, Pühringer D, Zielasek J, Hoeflich A, Silani V, Wolf E, Kneitz S, Sommer C, Toyka KV, Sendtner M, 2015. Dysregulated IGFBP5 expression causes axon degeneration and motoneuron loss in diabetic neuropathy. Acta Neuropathol. 130, 373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler RA, Gan Y, Zhang W, Liou AK, Gao Y, Cao G, Chen J, 2010. Heat shock proteins: cellular and molecular mechanisms in the central nervous system. Prog.Neurobiol 92, 184–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yuan H, Shu L, Xie J, Zhang D, 2004. Prolongation of T(2) relaxation times of hippocampus and amygdala in Alzheimer’s disease. Neurosci.Lett 363, 150–153. [DOI] [PubMed] [Google Scholar]
- Williamson W, Lewandowski AJ, Forkert ND, al, e., 2018. Association of cardiovascular risk factors with mri indices of cerebrovascular structure and function and white matter hyperintensities in young adults. JAMA. 320, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MP, Hugge C, Bielinska M, Nicholas P, Majerus PW, Wilson DB, 2009. Neural tube defects in mice with reduced levels of inositol 1,3,4-trisphosphate 5/6-kinase. Proc.Natl.Acad.Sci.U.S.A 106, 9831–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Vandal AC, Friston KJ, Evans AC, 1996. A unified statistical approach for determining significant signals in images of cerebral activation. Hum.Brain Mapp 4, 58–73. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Sato M, Maeda M, Takagi H, 1998. Identification of a novel adenylate kinase system in the brain: cloning of the fourth adenylate kinase. [DOI] [PubMed]
- Yu L, Boyle PA, Nag S, Leurgans S, Buchman AS, Wilson RS, Arvanitakis Z, Farfel JM, De Jager PL, Bennett DA, Schneider JA, 2015. APOE and cerebral amyloid angiopathy in community-dwelling older persons. Neurobiol.Aging 36, 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Dawe RJ, Boyle PA, Gaiteri C, Yang J, Buchman AS, Schneider JA, Arfanakis K, De Jager PL, Bennett DA, 2017. Association Between Brain Gene Expression, DNA Methylation, and Alteration of Ex Vivo Magnetic Resonance Imaging Transverse Relaxation in Late-Life Cognitive Decline. JAMA Neurol. 74, 1473–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, De Jager PL, Yang J, Trojanowski JQ, Bennett DA, Schneider JA, 2015. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 84, 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Petyuk VA, Gaiteri C, Mostafavi S, Young-Pearse T, Shah RC, Buchman AS, Schneider JA, Piehowski PD, Sontag RL, Fillmore TL, Shi T, Smith RD, De Jager PL, Bennett DA, 2018. Targeted brain proteomics uncover multiple pathways to Alzheimer’s dementia. Ann.Neurol [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.