Figure 6.
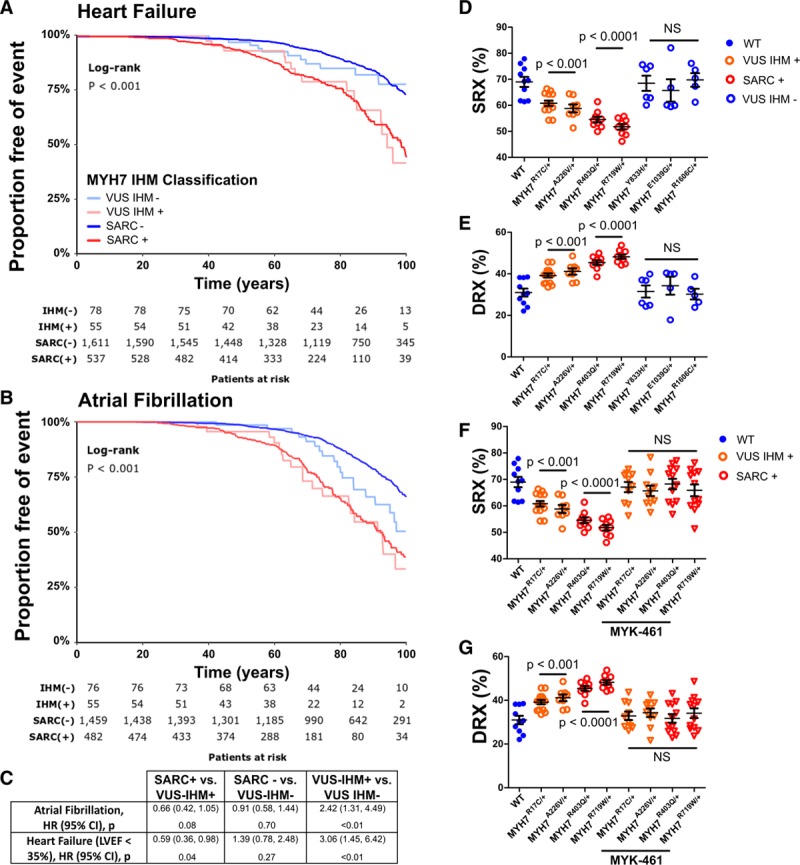
Myosin VUS that alter IHM residues are associated with higher rates of adverse clinical outcomes. A, Kaplan-Meier curves show relative time points at which SHaRe subjects, grouped according to genotype, meet composite heart failure end points (first occurrence of cardiac transplantation, LV assist device implantation, LV ejection fraction <35%, or New York Heart Association class III/IV symptoms). Genotypes are denoted as pathogenic sarcomere variants (SARC+), no pathogenic sarcomere variants (SARC–), variant of unknown significance that alters an IHM residue (VUS–IHM+), and variant of unknown significance that does not alter an IHM residue (VUS–IHM–). B, Kaplan-Meier curves demonstrate time to atrial fibrillation among SHaRe subjects with genotypes SARC+, SARC–, VUS–IHM+, and VUS–IHM–. (See also Figure IV in the online-only Data Supplement). C, Cox proportional hazard ratios comparing clinical SHaRe subjects with genotypes SARC+, SARC–, VUS–IHM+, and VUS–IHM– and adverse clinical outcomes of heart failure and atrial fibrillation. Significance was calculated using the log-rank test. D and E, Proportion of myosins in the SRX (D) and DRX conformations (E) in human ventricular tissues from healthy donor tissues (WT) or from HCM subjects with genotypes SARC+, VUS IHM+, (SARC–), and VUS IHM–. F and G, Proportion of myosin heads in SRX (F) and DRX (G) from healthy human ventricular tissues (WT) or HCM hearts with genotypes SARC+, VUS IHM+, (SARC–), and VUS IHM– region (VUS IHM +) before or after treatment with 0.3 μmol/L MYK461. All data are displayed with mean±SEM, and all significances are reported with a significance cutoff of P<0.05 after using two-way ANOVA with Bonferroni corrections for multiple comparisons. DRX indicates disordered relaxed state conformation of myosin molecule; HCM, hypertrophic cardiomyopathy; HR, hazard ratio; IHM, interacting-heads motif; LV, left ventricular; LVEF, left ventricular ejection fraction; NS, not significant; SHaRe, Sarcomeric Human Cardiomyopathy Registry; and SRX, super relaxed state conformation of myosin molecule.
