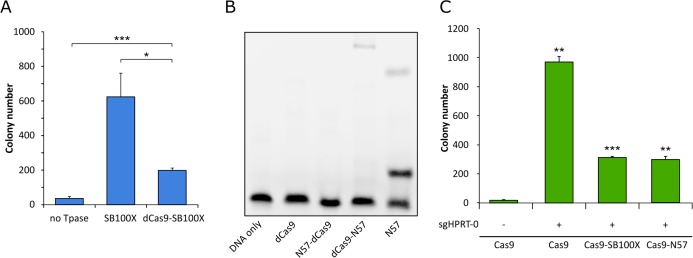
Figure 4. Functional testing of dCas9 fusions.
(A) Numbers of puromycin-resistant colonies in the transposition assay. The dCas9-SB100X fusion protein catalyzes ~30% as many integration events as unfused SB100X transposase (n = 3, biological replicates, *p≤0.05, ***p≤0.001, error bars represent SEM). (B) EMSA with dCas9-N57 fusion proteins. dCas9 serves as negative control, N57 as positive control. Binding can be detected for dCas9-N57, but not for N57-dCas9. The upper band in the positive control lane is likely a multimeric complex of DNA-bound N57 molecules, in line with N57’s documented activity in mediating protein-protein interaction between transposase subunits and in forming higher-order complexes (Izsvák et al., 2002). (C) Numbers of 6-TG resistant colonies after Cas9 cleavage. No disruption of the HPRT gene, as measured by 6-TG resistance over background, can be detected without the addition of an sgRNA. In the presence of sgHPRT-0, all Cas9 constructs cause significant disruption of the HPRT gene (n = 3, biological replicates, **p≤0.01, ***p≤0.001, error bars represent SEM).