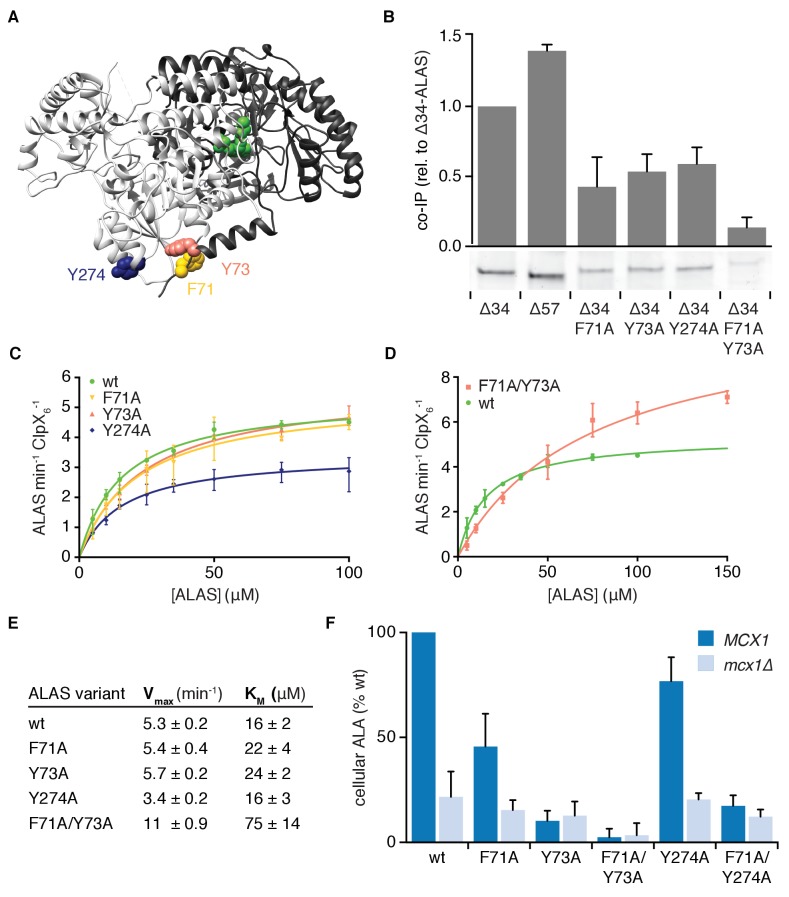
Figure 3. Multiple sequence-specific contacts direct mtClpX action on ALAS.
(A). The position of mutations that perturb mtClpX binding are mapped on one face of the structure of ALAS (PDB: 5TXR [Brown et al., 2018]) (F71 in yellow, Y73 in orange, and Y274 in dark blue). PLP is depicted in green. (B) Coimmunoprecipitation of ALAS variants with mtClpXE206Q-3xFLAG. 1 μM ALAS (monomer) and 0.5 μM mtClpXEQ-3xFLAG (hexamer) were incubated on ice with anti-FLAG antibody-conjugated magnetic beads. Coprecipitating proteins were eluted with 3xFLAG peptide. Eluted proteins were separated by SDS-PAGE and stained with Sypro Red. The bar graph above each lane of the gel represents the average intensity of the band from three independent experiments, normalized to wildtype Δ34-ALAS; error bars represent SD. (C–D) The rates at which mtClpX stimulated PLP binding to indicated ALAS variants are plotted as a function of ALAS monomer concentration; rates were determined and fit to the Michaelis-Menten equation as in Figure 2C. mtClpX was present at 0.5 μM hexamer (C) or 1 μM hexamer (D). Wildtype ALAS (Δ34-ALAS) data represented in Figure 2C is replotted in both panels. N = 3 for all variants. (E) Chart of parameters extracted from fits in (C–D) as in Figure 2C. Standard error of the fit is stated. (F) The levels of ALA in extracts from yeast strains harboring the indicated mutations in ALAS (HEM1 gene), with (MCX1) or without (mcx1Δ) the gene encoding mtClpX, were measured by colorimetric assay with modified Ehrlich’s reagent. p<0.001 for reduced ALA production in MCX1 strains by all mutations displayed in HEM1; p=0.05 for reduced ALA production in mcx1Δ hem1F71A/Y73A, n ≥ 3 for all strains; error bars represent SD.