Figure 1. Structural modeling of MCUB and the mitochondrial calcium uniporter (mtCU).
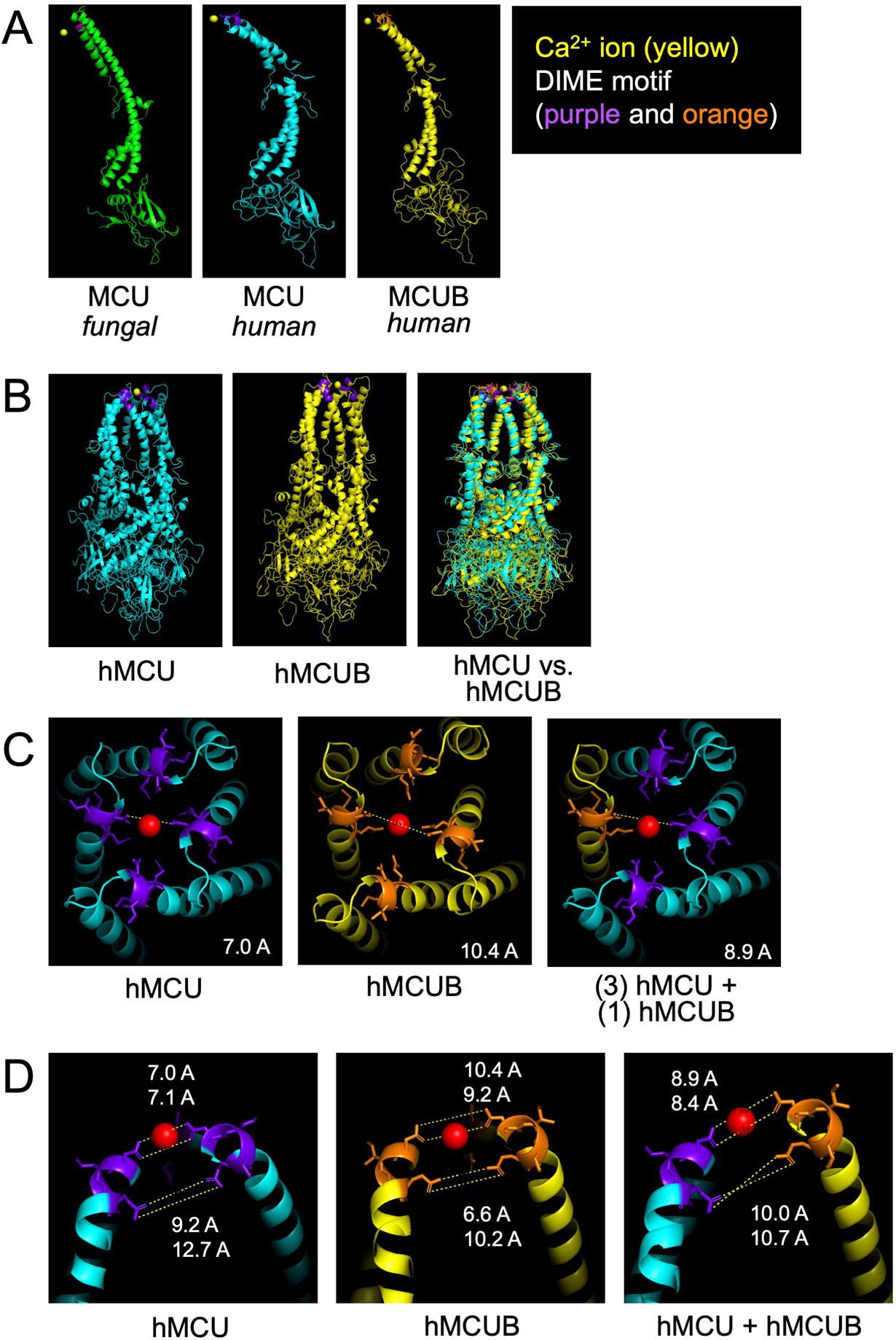
Using Modeller36, a program for comparative three dimensional protein structure analysis38, we uploaded the structural data for fungal MCU (PDB 6D7W) reported in Fan et al.37 and predicted the structure of human MCU followed by subsequent prediction of human MCUB. A) Comparison of monomeric structure. B) Comparison of human MCU and MCUB homo-oligomeric tetrameric structure. C) Zoom view of the mouth of the hydrophilic ionic pore formed by MCU tetramer, MCUB homo-oligomeric tetramer and a hetero-oligomeric tetramer (1 MCUB : 3 MCU). Red sphere represents a calcium (Ca2+) ion and purple and orange highlight the DIME motif. The dashed white line is a theoretic measurement in angstroms (A) between Aspartate-261 MCU residues or Aspartate-322 MCUB residues. D) Zoom of the sideview of opposing DIME motifs of MCU, MCUB, and MCU/MCUB hetero-oligomeric structure.
