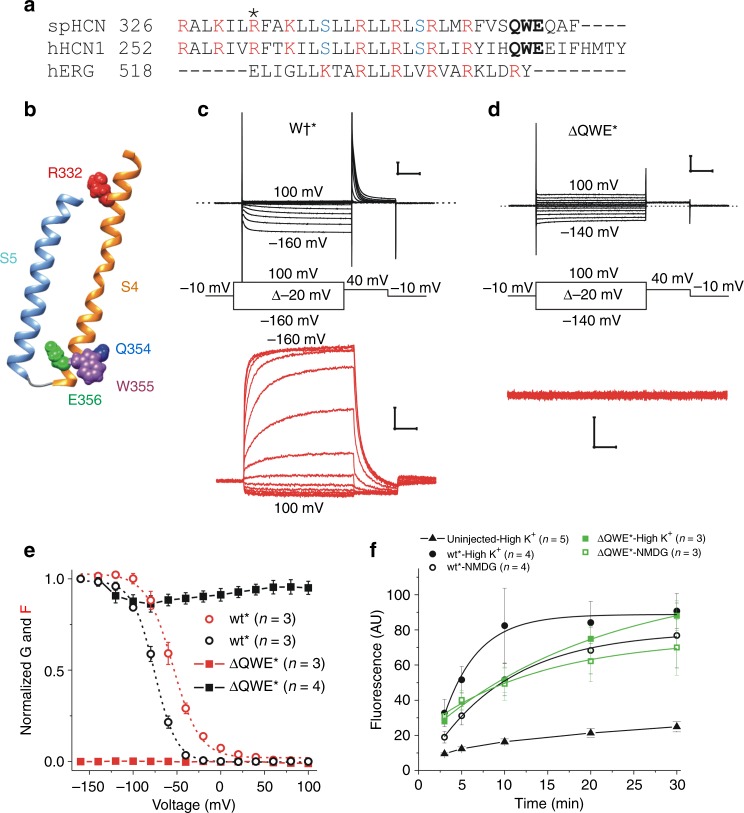
Fig. 1. Deleting QWE at the C-terminal S4 leads to an always open channel with an immobilized S4.
a S4 sequence alignment of spHCN, hHCN1, and hERG channels. The conserved QWE sequence is shown in bold. R332 is shown with an *. b spHCN homology model showing the position of the QWE residues. c, Representative traces of the current (black) and fluorescence (red) from c wt* and d ΔQWE* channels. Scale bars: 0.1 s, 5 µA, and 2% ΔF/F. e Normalized G(V) (black) and F(V) (red) curves of wt* and ΔQWE* channels. f Tetramethyl-rhodamine C5-maleimide labeling of uninjected oocytes and oocytes expressing the R332C (wt) and R332C/ΔQWE (ΔQWE) channels in high K+ and NMDG solutions. Labeling rates (k): 0.25 ± 0.06 (wt in high K+), 0.11 ± 0.01 (p = 0.05) (wt in NMDG), 0.05 ± 0.01 (p = 0.01) (ΔQWE in high K+), 0.07 ± 0.02 (p = 0.001) (ΔQWE in NMDG). Mean ± SEM. p values indicate statistical difference from wt in high K+ solution. Source data are provided as a Source Data file.