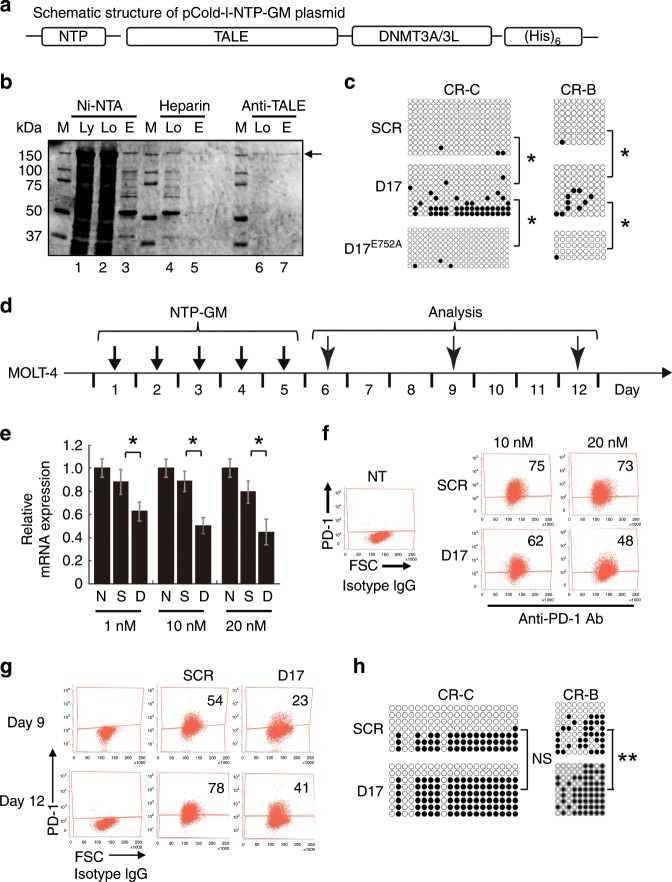
Fig. 2. Large-scale preparation of NTP-GM-D17 and effects of NTP-GM-D17 on PD-1 expression and DNA methylation of the PD-1 promoter region.
a Schematic structure of pCold-I-NTP-GM. The vector contained NTP, TALE, DNMT3A/3L and a (His)6 tag for purification. b CBB staining patterns of purification of NTP-GM-D17 protein. The arrow indicates purified NTP-GM-D17. M, marker; Ly, initial lysate; Lo, loaded sample; E, elution. c Bisulfite sequence analysis of CR-C and CR-B. Purified NTP-GM-D17 was subjected to an in vitro DNA methylation assay. *P < 0.01. d Experimental protocol for the treatment of MOLT-4 cells with NTP-GM-D17. After 5 days of treatment with NTP-GM-D17, MOLT-4 cells were cultured for another 7 days without NTP-GM-D17. On days 6, 9 and 12 after the initial addition of the protein, the cells were harvested and subjected to analysis. e NTP-GM-D17 reduces the expression of PD-1 mRNA. MOLT-4 cells were treated for 5 days with 1, 10 and 20 nM NTP-GM-D17. On day 6 after the initial addition of the protein, the cells were harvested and subjected to quantitative real-time PCR analysis. PD-1 mRNA expression levels were normalised to the level of GAPDH mRNA. Data are shown as the mean ± SD (n = 3). *P < 0.01. f FACS analysis of PD-1+ MOLT-4 cells. Treatment with 10 or 20 nM NTP-GM-D17 reduced the number of PD-1+ cells. Each number indicates the percentage of PD-1+ cells. g Dot plot of one representative result of FACS analysis of PD-1+ MOLT-4 cells. Cells were treated for 5 days with 20 nM NTP-GM-D17 and harvested on days 9 and 12. h NTP-GM-D17 induced DNA methylation of the PD-1 promoter region that persisted for 7 days. Data from bisulfite sequence analysis of CR-C and CR-B are shown. MOLT-4 cells treated for 5 days with NTP-GM-D17 and harvested on day 12. **P < 0.05; NS, not significant.