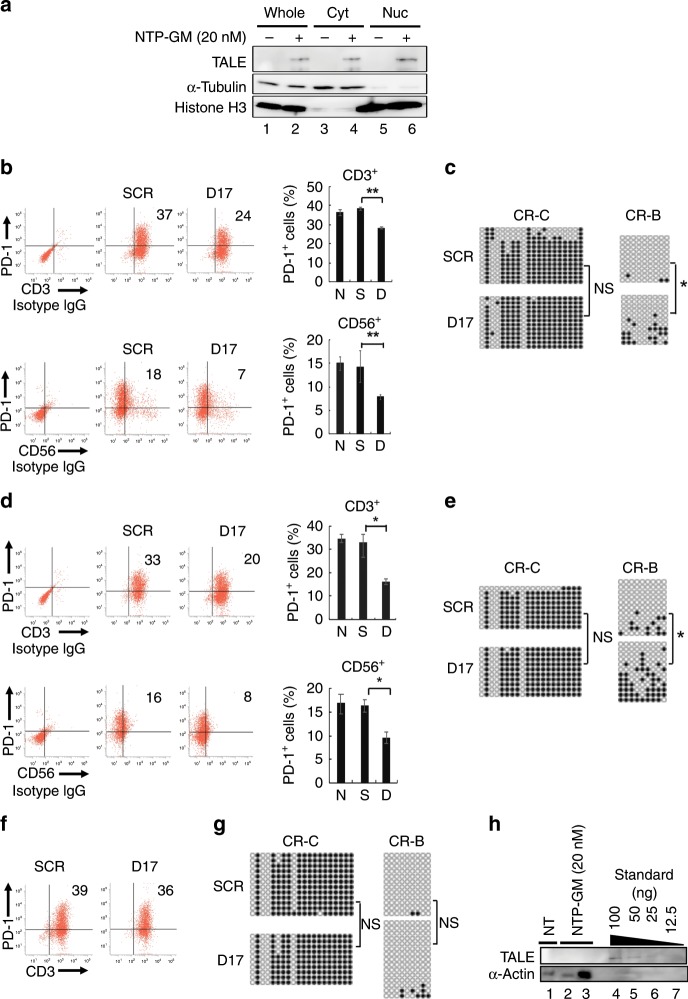
Fig. 3. Effects of NTP-GM-D17 on PBMCs.
a NTP-GM protein is translocated to the nucleus. PBMCs (1.0 × 107 cells) were treated with 20 nM NTP-GM-D17 for 12 h and cellular fractionation was performed. Anti-α-tubulin and anti-histone H3 antibodies were used for detecting proteins localised in cytosolic or nuclear fractions, respectively. Whole, whole lysate. Cyt, cytoplasmic fraction. Nuc, nuclear fraction. b NTP-GM-D17 reduces the number of PD-1+/CD3+ and PD-1+/CD56+ cells. Dot plot from one representative FACS analysis (left) and a graph showing integrated data from three independent experiments (right) are shown. Values represent the mean ± SD (n = 3). c Bisulfite sequence analysis of the PD-1 promoter region. Results for CR-C and CR-B in PBMCs, which were harvested on day 6 (see Fig. 2d) are shown. DNA methylation of CR-B was significantly increased by NTP-GM-D17. *P < 0.01; NS, not significant. d NTP-GM-D17 reduces the number of PD-1+/CD3+ and PD-1+/CD56+ cells on day 9 (see an experimental protocol shown in Fig. 2d). A graph of integrated data from three independent experiments was shown in the right panel. Values represent the mean ± SD (n = 3). e DNA methylation persists for 4 days after the last addition of NTP-GM-D17. Bisulfite sequence analysis of CR-C and CR-B was performed using PBMCs that were harvested on day 9. *P < 0.01; NS, not significant. f Effects of NTP-GM-D17 were cancelled on day 12. A representative result was shown. g Increased DNA methylation was restored on day 12. Data of bisulfite sequence analysis done on day 12 are depicted. NS, not significant. h Incorporated NTP–GP protein is degraded quickly. After treatment of PBMCs with 20 nM NTP-GM protein for 5 days, the cells were harvested on the following day, and whole-cell extracts of 1.0 or 5.0 × 105 cells (lanes 2 and 3) were subjected to western blot analysis. As control, 100 (lane 4), 50 (lane 5), 25 (lane 6) and 12.5 ng (lane 7) of NTP-GM protein was loaded onto the same SDS-PAGE gel.