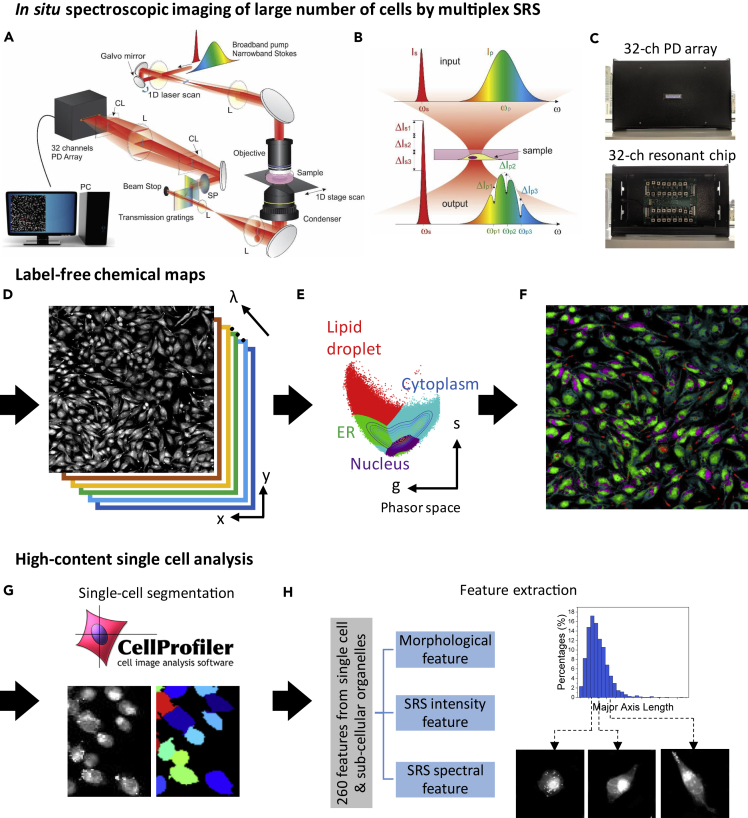
Figure 1.
Workflow of the Single-Cell Analysis by Multiplex SRS Imaging Cytometry
(A–C) In situ spectroscopic imaging of a large number of cells by multiplex SRS. (A) Hybrid scanning was implemented by scanning a galvo mirror while moving the motorized stage. Multiple Raman modes are parallelly detected by a laboratory-built 32-channel lock-in free resonant photodiode array detector. (B) Multiple Raman shifts are excited by a broadband pump beam and a narrowband Stokes beam. (C) A photograph of our laboratory-built 32-channel lock-in free resonant photodiode (PD) array detector. Upper panel: the detector front side, showing a 32-channel PD array. Lower panel: the detector backside, showing 32-channel resonant circuit chips for lock-in free detection.
(D–F) Label-free chemical mapping. (D) x-y-λ, a three-dimensional dataset generated by the multiplex SRS imaging cytometer. (E) The SRS spectrum from each image pixel is projected onto a 2D phasor domain, followed by an unsupervised clustering algorithm to separate the ER, nuclei, cytosol, and LDs. (F) A chemical map is generated by remapping the clustered results in the phasor domain back to the SRS image.
(G and H) High-content single-cell analysis. (G) Single-cell segmentation by CellProfiler. (H) Left panel: a total of 260 features in each cell are extracted, which can be classified into morphological features, SRS intensity features, and SRS spectral features. Right: statistical analysis of each feature demonstrates cellular heterogeneity.