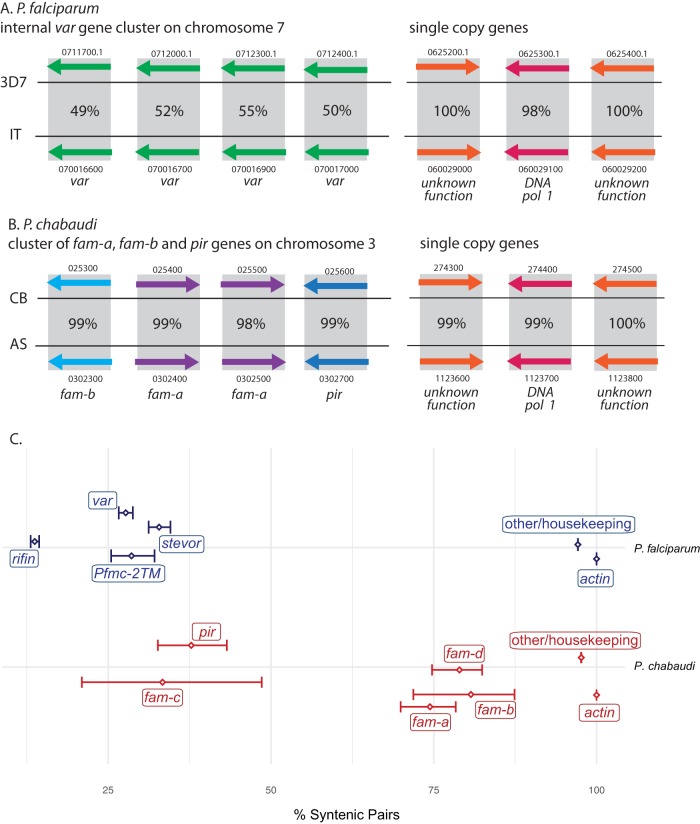
FIG 1.
Comparison of variant antigen diversity in human and rodent malaria parasites. (A) (Left) Schematically shown are four members of the multicopy variant antigen var gene family of the human parasite P. falciparum. Genes from two geographical isolates (3D7 and IT) are shown from a syntenic region of chromosome 7, and the percentage of nucleotide identity between each gene is provided in the gray box enclosing each gene pair. Annotation numbers corresponding to the Eukaryotic Pathogen Genomics Database Resource (Release 45, EuPathDB, eupathdb.org [35]) are included above each arrow. (Right) A similar schematic shows the near-complete sequence identity observed for single-copy housekeeping genes. (B) A similar analysis as shown in panel A for two isolates (CB and AS) of the rodent parasite P. chabaudi. (Left) Members of the variant gene families fam-a, fam-b, and pir. (Right) Single-copy housekeeping genes. Sequence identities were calculated using Needleman-Wunsch alignment of two sequences (36). (C) Assessment of recombination within the multigene families. Individual genes from 15 independent isolates of P. falciparum (top, blue text) were compared to the 3D7 reference genome to identify the ortholog with the highest-scoring sequence alignment. For single-copy housekeeping genes, isolate-to-reference gene pairs were in the syntenic position of the genome nearly 100% of the time (right); such pairs for members of the var, rifin, stevor, and Pfmc-2TM variant gene families were seldom syntenic (left), indicating extensive recombination throughout these families. Similarly, 4 isolates of the rodent malaria parasite P. chabaudi (bottom, red) were compared to the AS reference genome and demonstrated that a large majority of the fam-a, fam-b, and fam-d multigene family members maintained synteny, even across two subspecies. Diamonds represent percent synteny with 95% confidence intervals shown by error bars. See Text S1 in the supplemental material for details of sequence analysis.