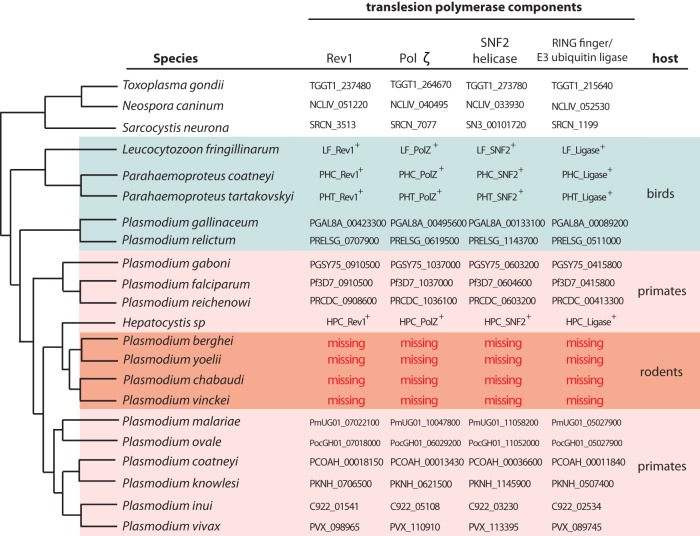
FIG 2.
Phylogenetic tree of apicomplexan parasites based on the phylogeny of Galen et al. (37) showing the loss of translesion polymerases in different parasite lineages. Toxoplasma gondii, Neospora caninum, and Sarcocystis neurona are parasites that do not possess large multigene families and do not undergo antigenic variation. Clades of parasites that infect red blood cells are highlighted with different-colored shading according to their vertebrate hosts. Rev1, Pol ζ, an SNF2 helicase, and a RING finger/E3 ubiquitin ligase are required for translesion polymerase activity. This analysis shows the loss of translesion polymerases in parasites that infect rodents. Gene annotation numbers are provided next to each species name for all species catalogued in the EuPathDB database (Release 45, EuPathDB, eupathdb.org [35]). Additional orthologous sequences were obtained from the fragmented genome assembly of Parahaemoproteus tartakovskyi (38), the transcriptome data sets of Parahaemoproteus coatneyi and Leucocytozoon fringillinarum (39), and sequence data of a Hepatocystis parasite that were mined from the transcriptome of a Ugandan red colobus monkey (40) using the ContamFinder pipeline (41). +, see Fig. S1 in the supplemental material for sequences and alignments of genes not previously annotated.