Abstract
Tyrosine kinase inhibitors (TKIs) induce autophagy in many types of cancer cells. We previously reported that gefitinib (GEF) and imatinib (IMA) induce autophagy in epidermal growth factor receptor (EGFR) knock-out A549 and non-BCR-ABL-expressing leukemia cell lines, respectively. This evidence suggests that TKI-induced autophagy is independent of the original target molecules. The present study compared the autophagy-inducing abilities of various TKIs, regardless of their targets, by quantitative autophagy flux assay. We established stable clones expressing the GFP-LC3-mCherry-LC3ΔG plasmid in A549, PC-9, and CAL 27 cell lines and assessed autophagy inducibility by monitoring the fluorescent ratios of GFP-LC3 to mCherry-LC3ΔG using an IncuCyte live cell imaging system during exposure to TKIs viz; GEF, osimertinib (OSI), lapatinib (LAP), lenvatinib (LEN), sorafenib (SOR), IMA, dasatinib (DAS), and tivantinib (TIV). Among these TKIs, DAS, GEF, and SOR exhibited prominent autophagy induction in A549 and PC-9 cells. In CAL 27 cells, IMA, SOR, and LEN, but not GEF, TIV, or OSI, exhibited autophagy induction. In the presence of azithromycin (AZM), which showed an inhibitory effect on autophagy flux, TKIs with prominent autophagy inducibility exhibited enhanced cytotoxicity via non-apoptotic cell death relative to effects of TKI alone. Therefore, autophagy inducibility of TKIs differed in the context of cancer cells. However, once induced, they appeared to have cytoprotective functions. Thus, blocking TKI-induced autophagy with AZM may improve the therapeutic effect of TKIs in cancer cells.
Keywords: Tyrosine kinase inhibitor, Autophagy, Macrolide antibiotics, Cancer
Abbreviations: tyrosine kinase inhibitors, TKIs; gefitinib, GEF; imatinib, IMA; osimertinib, OSI; lapatinib, LAP; lenvatinib, LEN; sorafenib, SOR; dasatinib, DAS; tivantinib, TIV; azithromycin, AZM; bafilomycin A1, BAF; receptor tyrosine kinase, RTK
Highlights
-
•
Tyrosine kinase inhibitors (TKIs) induce autophagy regardless of their main target.
-
•
This autophagy inducibility is partially determined in the context of cancer cells.
-
•
Azithromycin (AZM) has an inhibitory effect on autophagy.
-
•
Blocking TKI-induced autophagy with AZM enhances their cytotoxicity in cancer cells.
-
•
This enhanced cytotoxicity is mediated through non-apoptotic cell death.
1. Introduction
Autophagy is a cellular self-digestive system in eukaryotes that maintains the cellular homeostasis by recycling the intracellular components. Autophagy is responsible for mitochondrial turnover and removal of damaged mitochondria producing reactive oxygen species. Thus, autophagy prevents tumorigenesis by elimination of the harmful mitochondria that could cause genotoxicity. However, once cancer has been established, autophagy appears to play a cytoprotective role for adaptation to hypoxic and lower nutrient environments due to insufficient vascularity and metastasis, in addition to therapeutic resistance. Thus, autophagy plays dual functional roles in tumorigenesis and its progression [1,2].
The treatment with tyrosine kinase inhibitors (TKIs) including epidermal growth factor receptor (EGFR)-TKIs induces autophagy in various types of cancer cells [[3], [4], [5]]. In the molecular mechanism of autophagy induction in response to TKIs, activation of receptor tyrosine kinase (RTK) itself potently inhibits autophagy by activating the PI3K-AKT-mTOR pathway, one of the downstream signaling pathways of RTKs. This often occurs in many cancer cells with overexpressed and/or mutated EGFR family of proteins including non-small cell lung cancer (NSCLC) and breast cancer [6]. The mTOR activation negatively regulates autophagy through the inhibition of unc-51-like kinase 1 (ULK1) complex as well as repression of transcription factor E (TFE) and autophagy-related (ATG) genes through mTOR-dependent phosphorylation [7,8]. Furthermore, active EGFR binds Beclin 1 leading to its multisite tyrosine phosphorylation, enhanced binding to the autophagy inhibitor Rubicon, and decreased Beclin 1-associated VPS34 (PI3 kinase) activity, resulting in the inhibition of autophagosome formation [9]. Therefore, treatment with EGFR-TKIs, such as gefitinib (GEF) or erlotinib, appears to release autophagy repression from the multi-regulatory mechanisms.
We have previously reported that GEF treatment induced autophagy in both EGFR knockout-A549 and leukemia cell lines derived from the mesoderm without EGFR expression [10]. We had also reported that imatinib mesylate (IMA) treatment induced autophagy in various cancer cell lines without BCR-ABL expression [11]. These data strongly suggested that TKI-induced autophagy is independent of their original target molecule(s). In this context, other target(s) for autophagy regulatory molecule(s) in response to TKI treatment might exist [10,11]. In contrast to autophagy induction by treatment with TKIs, we have reported the potent inhibitory effect of autophagy flux induced by treatment with macrolide antibiotics including azithromycin (AZM) and clarithromycin [12,13]. Although treatment with macrolide antibiotics alone showed almost no cytotoxicity, combined treatment with GEF and macrolide resulted in enhanced cytotoxicity of GEF in NSCLC cell lines [10]. In pancreatic cancer cell lines, this pronounced cytotoxicity of EGFR-TKIs appeared to depend on the autophagy inhibitory efficacy of macrolides, with AZM being the most potent autophagy inhibitor among the macrolides tested, and it exhibited the most prominent enhanced cytotoxicity in combination with GEF [14]. In contrast, the question has been raised whether the autophagy inducibility of TKIs contributes to enhanced cytotoxicity when their cytoprotective autophagy is blocked by AZM.
The present study compared the autophagy inducibility of TKIs regardless of their original target by quantitative autophagy flux assay using the GFP-LC3-mCherry-LC3ΔG autophagy flux probe [15]. We further examined whether blocking TKI-induced-autophagy using AZM further enhanced their cytotoxic effects.
2. Materials and methods
2.1. Reagents
GEF, lapatinib (LAP), tivantinib (TIV), and sorafenib (SOR) were purchased from Cayman Chemical Company (Ann Arbor, MI, USA). Dasatinib (DAS) was purchased from ChemScene (Monmouth Junction, NJ, USA) and imatinib (IMA) and AZM were obtained from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Osimertinib (OSI) was purchased from Toronto Research Chemicals Inc (North York, Canada), and lenvatinib (LEN) from LC Laboratory (Woburn, MA, USA). Bafilomycin A1 (BAF) and Hank’s Balanced Salt Solution (HBSS) were obtained from Fujifilm Wako Pure Chemicals Corporation (Osaka, Japan).
2.2. Cell line and culture conditions
An NSCLC cell line PC-9 harboring EGFR mutations was obtained from RIKEN Bio Resource Center (Tsukuba, Japan). The NSCLC cell line A549 and a human oral squamous cell carcinoma cell line CAL 27, both having wild-type EGFR, were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) [16]. The A549 and PC-9 cells were cultured in RPMI-1640 medium (Sigma-Aldrich, Merck, St. Louis, MO, USA), while the CAL 27 cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) with high glucose (Sigma-Aldrich, Merck) supplemented with 10% fetal bovine serum (FBS, Biosera, Ringmer, UK) and 1% Penicillin-Streptomycin solution (Fujifilm Wako Pure Chemical Industries) in a humidified incubator containing 5% CO2 at 37 °C. The cells were cultured for up to one month after thawing. Mycoplasma contamination was routinely assayed using an e-MycoTM Mycoplasma PCR Detection Kit (iNtRON Biotechnology, Inc., Korea). As starvation culture for autophagy induction, the cells were cultured with HBSS without FBS for the indicated time period.
2.3. Assessment of the numbers of viable and dead cells
The cells were pre-seeded in a 96-well plate for 24 h to allow them to adhere to the flat bottom of the culture plate; subsequently, various TKIs were added to each well. The number of viable cells was assessed using CellTiter-Blue assay kit (Promega, Madison, WI) as previously reported [13]. The number of dead cells were assessed by staining with propidium iodide (PI) (Wako Pure Chemicals Corporation); the red fluorescence signal was monitored using the IncuCyte ZOOM (Essen Bioscience, Ann Arbor, MI) automated live cell imaging system.
2.4. Immunoblotting
Immunoblotting was performed as previously reported [13]. The transferred membranes were probed with the following primary polyclonal or monoclonal (m) antibodies (Abs): anti-p62/SQSTM1 (sc-28359), anti-GFP (sc-9996), and anti-β-actin (C4) mAbs (sc-47778) from Santa Cruz Biotechnology (Dallas, TX, USA); anti-poly ADP-ribose polymerase (PARP) Ab (#9542S) and anti-caspase-3 Ab (#9662) from Cell Signaling Technologies (Danvers, MA, USA); anti-LC3B Ab (NB600-1384) from Novus Biologicals (Littleton, CO); and anti-mCherry Ab (ab167453) from Abcam plc (Cambridge, UK). After incubation with the appropriate secondary peroxidase-conjugated Ab for 1–2 h, the immune-reactive proteins were detected using the Immobilon Western Chemiluminescent HRP Substrate (WBKLS0500, Millipore/Merck, Burlington, MA, USA). Densitometry was performed using a WSE-6300H/C LuminoGraph III (ATTO, Tokyo, Japan).
2.5. Morphological assessment
After treatment with trypsin, the cells were spread on the glass slides using a Cytospin 4 centrifuge (Thermo Fisher Scientific, Waltham, MA, USA) to make the glass slide preparations that were stained with May-Grünwald-Giemsa. The morphological observations were performed using a digital microscope (BZ-8100, Keyence Co., Osaka, Japan).
2.6. Establishment of GFP-LC3-mCherry-LC3ΔG-expressing cell lines
The pMRX-IP-GFP-LC3-RFP-LC3ΔG plasmid was a kind gift from Professor N. Mizushima (University of Tokyo, Tokyo, Japan) [15]. RFP was replaced with mCherry to obtain a higher fluorescence intensity as follows: the plasmid without the RFP region was linearized by polymerase chain reaction (PCR) using the KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA, USA) and the primers 5’-GGCCGCCACTCCACCGGCGCC-3’ and 5’-CCCGAACGTCTCCTGGGAGGC-3’. The mCherry region of the mCherry2-C1 plasmid (#54563, Addgene, Cambridge, MA, USA) was amplified by PCR using the same kit and primers 5’-CAGGAGACGTTCGGGATGGTGAGCAAGGGCGAGGAG-3’ and 5’-GGTGGAGTGGCGGCCCTTGTACAGCTCGTCCATGCC-3’. These fragments were combined using the In-Fusion HD Cloning Kit (Takara Bio, Shiga, Japan). The resultant plasmid, pMRX-IP-GFP-LC3-mCherry-LC3ΔG, was confirmed by sequence analysis. The stable cell lines were generated as follows: pMRX-IP-GFP-LC3-mCherry-LC3ΔG was electroporated into A549, PC-9, and CAL 27 cells using the NEPA21 Super Electroporator (Nepa Gene, Chiba, Japan). The transfected cells were selected with puromycin and single clones were isolated using the cloning ring. GFP-LC3 and mCherry-LC3ΔG expression was confirmed by fluorescence microscopy and immunoblotting using specific Abs against GFP and mCherry.
2.7. Statistical analysis
All data are expressed as mean ± SD. The statistical analysis included two-way analysis of variance (ANOVA), followed by Bonferroni multiple comparisons test using GraphPad Prism software. P-values < 0.05 were considered to indicate statistically significant differences.
3. Results
3.1. Quantitative autophagy flux assay using the GFP-LC3-mCherry-LC3ΔG probe and IncuCyte live cell imaging system
We first transfected pMRX-IP-GFP-LC3-mCherry-LC3ΔG into A549 cells and selected the stable expressing clone A549-GFP-LC3-mCherry-LC3ΔG with sufficient GFP and mCherry fluorescent intensities for the autophagy flux assay, as described in the Materials and methods section [15]. The cells were then cultured in either HBSS (amino acid-depleted condition) for autophagy induction or in the presence of 10 nM of BAF for blocking of autophagy flux. The fluorescent intensities of GFP and mCherry were continuously monitored using the IncuCyte live cell imaging system. Regarding the GFP-LC3-mCherry-LC3ΔG probe, LC3-I covalently links to phosphatidylethanolamine (PE) to form LC3-II, which is incorporated into the autophagosomal membrane. Thus, during autophagic processing, LC3-II on the inner side of the autophagosomal membrane is degraded upon fusion with the lysosome, whereas mCherry-LC3ΔG lacking the C-terminal glycine residue linking to PE was preserved as an internal control because it is not involved in the autophagosome [15]. As shown in Fig. 1A, autophagy induction under HBSS culture conditions resulted in the elimination of GFP-LC3 due to lysosomal degradation. In contrast, in the presence of BAF, prominent cytoplasmic accumulation of GFP-LC3 occurred when autophagy flux was blocked (Fig. 1A). During autophagy induction under HBSS culture, elimination of GFP-LC3 resulted in smaller GFP/mCherry ratios as compared to those cells that were cultured with complete culture medium, showing a downward curve when plotted. In contrast, the accumulation of GFP-LC3 due to blocking of the autophagy flux in the presence of BAF resulted in an upward curve (Fig. 1B). To validate this assay, we compared the immunoblotting results (Fig. 1C). Under HBSS culture conditions, the intensities of LC3B-II as an autophagosome marker increased along with almost complete elimination of LC3B–I during 12 h-culture with HBSS in A549-GFP-LC3-mCherry-LC3ΔG cells. Furthermore, the expression levels of p62, a substrate of autophagy, decreased within 12 h. In BAF-treated cells, the intensities of both LC3B-II and p62 bands increased continuously during the 24 h-exposure. These results well fitted the curves of the GFP/mCherry ratios as shown in Fig. 1B.
Fig. 1.
Validation of the quantitative autophagy flux assay using the GFP-LC3-mCherry-LC3ΔG probe by comparisons of the immunoblotting data in A549 cells.
A. A549-GFP-LC3-mCherry-LC3ΔG cells were cultured in Hank’s Balanced Salt Solution (HBSS) or complete culture medium with BAF (10 nM) for 12 h. DMEM high glucose supplemented with 10% FBS containing 1% penicillin/streptomycin solution and 0.3% DMSO was used as the control culture medium. The scale bar indicates 100 μm
B. Fluorescent ratios of GFP/mCherry in A549-GFP-LC3-mCherry-LC3ΔG cells were monitored every hour and plotted using an IncuCyte live cell imaging system during the 24 h BAF treatment or HBSS culture.
C. After treatment of A549-GFP-LC3-mCherry-LC3ΔG cells with BAF (10 nM) or HBSS culture for 12 h and 24 h, immunoblotting was performed using anti-LC3B, anti-p62, anti-GFP, and anti-mCherry Abs. Immunoblotting with anti-β-actin mAb was performed as an internal control.
3.2. Comparison of autophagy inducing ability of TKIs
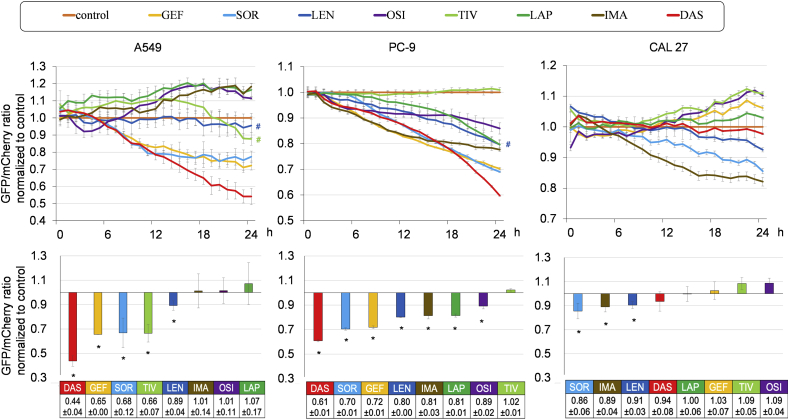
We next compared the autophagy-inducing abilities of various TKIs in the A549, PC-9, and CAL 27 cell lines stably expressing GFP-LC3-mCherry-LC3ΔG. The A549 and CAL 27 cells express wild-type EGFR, whereas PC-9 cells harbor EGFR-activating mutation [16]. We initially treated these cell lines with TKIs; namely, GEF, SOR, LEN, OSI, TIV, LAP, IMA, and DAS at fixed concentrations of 10 μM to compare their autophagy-inducing abilities. These TKIs exhibited different cytotoxicities for each cell line (Supplementary Fig. S1). Additionally, cells undergoing cell death in response to TKI treatment exhibited high transient auto-fluorescence intensity, causing incorrect evaluation of the autophagy inducibility (Supplementary Fig. S2). Thus, we treated the cells with TKIs at IC50 concentration at 48 h and assessed the GFP/mCherry ratios within 24 h-exposure to TKIs to exclude the influence of auto-fluorescence caused by cell death (Supplementary Table S1).
As shown in Fig. 2, the autophagy inducibilities of the TKIs at their IC50 values were evaluated and compared to those in the cells cultured in complete culture media at each time point. In A549 cells, five of eight TKIs tested exhibited autophagy induction whereas, in PC-9 cells, all but TIV exhibited autophagy induction. Notably, DAS (mainly targeting BCR-ABL), GEF (targeting EGFR), and SOR (multi-kinase inhibitor mainly targeting vascular endothelial growth factor receptor [VEGFR], SRC, and RAF) showed potent autophagy induction in both NSCLC cell lines. In CAL 27 cells, SOR and LEN (mainly targeting VEGFR) exhibited autophagy induction while GEF, TIV (targeting MET), and OSI (targeting EGFR) inhibited autophagy. IMA with the main target of BCR-ABL induced autophagy in PC-9 and CAL 27 cells but showed no apparent induction in A549 cells. These data suggest that TKIs induce autophagy regardless of their original main target(s) and that the autophagy inducibility of TKIs appeared to be determined in the context of cancer cells.
Fig. 2.
Assessment of the autophagy-inducing abilities of TKIs in A549, PC-9, and CAL 27 cells stably expressing the GFP-LC3-mCherry-LC3ΔG probe.
A549, PC-9, and CAL 27 cells stably expressing GFP-LC3-mCherry-LC3ΔG were treated with eight TKIs (gefitinib [GEF], sorafenib [SOR], lenvatinib [LEN], osimertinib [OSI], tivantinib [TIV], lapatinib [LAP], imatinib [IMA], and dasatinib [DAS]) at the IC50 concentrations for 48 h as shown in Supplementary Table S1. In case the cell growth inhibition did not reach 50% during the 48 h exposure time, the IC25 concentration at 24 h was used for the assessment of autophagy flux (#).
The upper panels plot the mean ± SD of GFP/mCherry ratios normalized to those of the cells cultured in control medium defined as 1.0 at each time point. Each upper panel is representative result of three independent experiments.
The lower panels represent the mean ± SD of the normalized GFP/mCherry ratios obtained from three independent experiments. *p<0.05, vs cells cultured with control medium containing 0.3% DMSO.
3.3. Blocking autophagy with AZM enhances the cytotoxic effect of TKIs with autophagy inducibility in cancer cell lines
We previously reported the autophagy inhibitory effects of macrolide antibiotics including AZM [12,13]. The A549-GFP-LC3-mCherry-LC3ΔG cells cultured with AZM exhibited the upward curve due to increased GFP/mCherry ratios, indicating autophagy inhibition (Fig. 3A).
Fig. 3.
Enhanced cytotoxicity of treatment with TKIs in combination with AZM through autophagy inhibition.
A. A549-GFP-LC3-mCherry-LC3ΔG cells were treated with AZM (5, 50, or 100 μM) for 24 h for autophagy flux assays, as shown in Fig. 2.
B. A549-GFP-LC3-mCherry-LC3ΔG cells were treated with control medium, SOR (13 μM; IC50), and AZM (50 μM), or the combination of SOR plus AZM at the same concentrations and processed for autophagy flux assays.
C. A549, PC-9, and CAL 27 cells were treated with TKIs (DAS, GEF, LEN, SOR, or OSI) +/- AZM (50 μM) for 48 h. Assessment of viable cell numbers was performed by CellTiter-Blue cell viability assay. (n=3) *p<0.05, **p<0.01.. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We then treated A549 and PC-9 cells with a combination of AZM plus either DAS, GEF, or SOR, which all exhibited potent autophagy induction (Fig. 2). This resulted in the pronounced cell growth inhibition of TKIs relative to the effects of each TKI alone (Fig. 3C). In CAL 27 cells, treatment with SOR or LEN induced autophagy and showed enhanced cytotoxicity in the presence of AZM. However, there was no increase in cytotoxicity for the combination of AZM and TKIs without autophagy inducibility, such as with GEF or OSI in CAL 27 cells (Figs. 2 and 3C). These data suggest that TKI-induced autophagy plays a cytoprotective role and that combined treatment with an autophagy inhibitor led to enhanced TKI cytotoxicity as long as autophagy was induced.
3.4. Non-apoptotic cell death induction by blocking TKI-inducing autophagy with AZM in cancer cell lines
To investigate whether the pronounced cell growth inhibition caused by treatment with a combination of TKI and AZM was due to apoptosis induction, we monitored cell death during 48 h-exposure to TKI and AZM. As shown in Fig. 4A, the number of PI-positive cells indicating dead cells became prominent at 24–48 h exposure to GEF and AZM in A549 and PC-9 cells. CAL 27 cells treated with SOR plus AZM showed a two-phase curve in cell death, with rapid induction within 6 h that was further pronounced after 24 h exposure. Surprisingly, we observed no PARP cleavage or caspase-3 activation in any of the three treated cell lines (Fig. 4B). The morphological assessment also indicated no typical features of cells undergoing apoptosis, such as chromatin condensation, nuclear fragmentation, and apoptotic body formation (Fig. 4C). The treated cells showed swollen cytoplasm and loss of plasma membrane integrity, which appeared to be consistent with necrosis or necroptosis [18].
Fig. 4.
Simultaneous treatment with TKI plus AZM enhances non-apoptotic cell death.
A. A549 and PC-9 cells (2.5 × 105 cells/mL) were treated with/without GEF (at IC50) +/- AZM (50 μM). CAL 27 cells were treated with/without SOR (6.0 μM; IC50) +/- AZM (50 μM). Dead, propidium iodide (PI)-stained cells were monitored using the IncuCyte system. The experiments were performed in triplicates and expressed as mean ± SD. Cells cultured in complete culture medium containing 0.3% DMSO were used as the control cells.
B. After treatment with TKIs (GEF for A549 and PC-9, SOR for CAL 27) ± AZM for 48 h, immunoblotting was performed using anti-PARP and anti-caspase-3 Abs. An A549-cell lysate treated with 10 μM staurosporine for 4 h was used as the positive control for apoptosis induction. Immunoblotting with anti-β-actin mAb was performed as an internal loading control.
C. Microscopic findings of cytospin preparations (after 48 h treatment) stained with May-Grünwald-Giemsa. Original magnification 1,000x. The scale bars represent 10 μm.
4. Discussion
Our previous reports indicated that TKI-induced autophagy was independent of the inhibition of the original target molecules, because GEF and IMA induced autophagy in EGFR knock-out A549 and non-BCR-ABL-expressing leukemia cell lines, respectively [10,11]. In the present study, we compared the autophagy inducibility of various TKIs for clinically targeting different kinase(s) by using quantitative autophagy flux assay. As shown in Fig. 2, seven out of eight TKIs that were tested exhibited significant autophagy induction in the NSCLC cell line PC-9 harboring activated EGFR mutation. In the NSCLC cell line A549 having the wild-type EGFR, treatment with GEF, SOR, and DAS exhibited potent autophagy inducibility; similar results were also seen in PC-9 cells with the EGFR mutation. However, in the human oral squamous cell carcinoma cell line CAL 27 having the wild-type EGFR, treatment with GEF and DAS did not exhibit significant autophagy induction. Therefore, the autophagy inducibility of TKIs appears to be determined in the context of cancer cells. Of note, SOR was a potent autophagy inducer in three cell lines tested. SOR is a multi-kinase inhibitor that simultaneously inhibits the RAF/MEK/ERK pathway as well as RTKs, such as VEGFR, platelet-derived growth receptor (PDGFR), and KIT [19]. Recent reports revealed the involvement of the RAF/MEK/ERK pathway in autophagy regulation independent of the PIK3/AKT/mTOR pathway [[20], [21]]. In B-RAFV600E-melanoma and B-RAFV600E-pancreatic ductal adenocarcinoma, autophagy was induced by the B-RAF inhibitor as part of the activation of lysosome biogenesis mediated by the upregulation of TFEB, which was phosphorylated and inactivated by B-RAFV600E via its downstream target ERK [[17], [21],22]. Inhibition of MEK1/2 also leads to the activation of the LKB1→AMPK→ULK1 signaling axis, a key regulator of autophagy [22]. Therefore, TKI in combination with the inhibitory effects of RAF/MEK/ERK pathway may exert more potent autophagy induction. Moreover, it was reported that SOR treatment induced autophagy via inhibition of SRC family kinases in gastrointestinal tumor cells [5]. In this regard, DAS, a multi-targeted inhibitor of ABL and SRC, as well as KIT and EGFR, also exhibited potent autophagy induction in the PC-9 and A549 cells (Fig. 2). Autophagy in response to SRC inhibition is accompanied by PI3K/AKT/mTOR signaling pathway inhibition, which shares the key regulatory cascade for autophagy in response to RTK-inhibition, as described above [6,22]. Furthermore, SRC-TKI induced autophagy depended on the induction of ULK1 kinase via down-regulation of microRNA-106a in lung carcinoma cells [23]. These authors reported that the ULK1 upregulation was mediated by the downregulation of the ULK1-targeting microRNA-106a in response to SRC-TKI, indicating the workings of the newly identified miR-106a-ULK1 signaling pathway [23]. Although other unidentified molecule(s) might participate in autophagy induction in response to TKIs, the total driving force via multi-regulatory pathways for autophagy including the 1) PIK3/AKT/mTOR pathway, 2) RAF/MEK/ERK cascade, and 3) SRC-miR-106a-ULK1 pathway might determine the autophagy inducibility of the TKIs.
To our surprise, combined treatment of TKI and AZM, which showed potent cytotoxicity in cancer cell lines, resulted in non-apoptotic cell death (Fig. 4). A recent study reported that the autophagy machinery serves as a scaffold for the activation of necroptosis signaling, which is independent of its self-digestive function [24]. Necroptosis requires the necrosome, a cytosolic complex formed by receptor-interacting serine/threonine-protein kinase 1 (RIPK1) in complex with RIPK3, Fas-associated protein with death domain (FADD), and caspase-8. Upon trans- and auto-phosphorylation of RIPK1/RIPK3, mixed kinase domain-like protein (MLKL) is recruited to the necrosome, phosphorylated, and ultimately mediates plasma membrane permeabilization for necroptosis by forming a cationic channel consisting of an MLKL octamer [25,26]. During this process, autophagosomes serve as a scaffold for the necrosome formation for the cells to undergo necroptosis [24]. The localization of the necrosome on the autophagosome appeared to require p62 binding to RIPK1 because the loss of p62 was sufficient to cause the switch of the type of cell death from necroptosis to apoptosis [24]. As shown in Fig. 3B, treatment with a combination of SOR and AZM led to the accumulation of intracellular autophagosomes, as indicated by an increased GFP/mCherry ratio, via blocking of autophagic flux at the late stages by AZM along with simultaneous autophagy induction by SOR. Therefore, localization of the necrosome complex to the accumulated cytoplasmic autophagosomes likely led to necroptosis rather than apoptosis. Thus, it is important to consider not just whether the autophagy pathway is inhibited but also at which point of the autophagy pathway the inhibition occurs.
The present study showed that many TKIs induced autophagy regardless of their original main target. However, the extent of autophagy induction appears to be determined in the context of cancer cells, as indicated by the different responses in three cell lines to treatment with the same TKI (Fig. 2). Thus, in the clinical setting, it is difficult to predict the potency of autophagy induction by TKI in various cancer cases for now. We have ongoing research focused at identifying the pivotal target molecule responsible for autophagy induction by TKIs, which may clearly determine the autophagy response in each case. However, as described above, the multi-kinase inhibitors appear to have a higher propensity for autophagy induction. Once autophagy was induced, blocking TKI-induced autophagy with AZM resulted in enhanced cytotoxicity via non-apoptotic cell death. These data suggested clinical benefit in cancer therapy for the combination therapy of TKI and AZM. However, further studies are required to elucidate the precise molecular mechanism of cell death and autophagy inhibition by AZM.
Funding
This study was supported by funds provided through the Strategic Research Foundation at Private Universities (S1411011, 2014–2018) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and by JSPS KAKENHI grant number JP17K08771 to KM.
CRediT authorship contribution statement
Hideki Tanaka: Investigation, Writing - review & editing. Hirotsugu Hino: Investigation, Methodology, Writing - review & editing. Shota Moriya: Methodology, Resources. Hiromi Kazama: Methodology, Investigation, Validation. Masaya Miyazaki: Investigation, Validation. Naoharu Takano: Investigation, Validation, Formal analysis, Writing - review & editing. Masaki Hiramoto: Validation, Writing - review & editing. Kiyoaki Tsukahara: Writing - review & editing, Supervision. Keisuke Miyazawa: Conceptualization, Supervision, Writing - review & editing, Funding acquisition.
Acknowledgements
We would like to thank Professor N. Mizushima (Univ. of Tokyo, Tokyo, Japan) for the kind gift of the pMRX-IP-GFP-LC3-RFP-LC3ΔG plasmid. We thank Editage (www.editage.com) for English language editing.
Footnotes
Transparency document related to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100750.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100750.
Transparency document
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.White E. Deconvolution the context-dependent role for autophagy in cancer. Nat. Rev. Canc. 2012;12:401–410. doi: 10.1038/nrc3262. https://doi:10.1038/nrc3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H.T., Liu H., Mao M.J. Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy. Mol. Canc. 2019;18:101. doi: 10.1186/s12943-019-1030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han W., Pan H., Chen Y. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cell. PloS One. 2011;6 doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Henson E.S., Xiao W. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy. 2016;12:1029–1046. doi: 10.1080/15548627.2016.1164357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park M.A., Reinehr R., Häussinger D. Sorafenib activates CD95 and promotes autophagy and cell death via Src family kinases in gastrointestinal tumor cells. Mol. Canc. Therapeut. 2010;8:2220–2231. doi: 10.1158/1535-7163.MCT-10-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henson E., Chen Y., Gibson S. EGFR Family members' regulation of autophagy is at a crossroads of cell survival and death in cancer. Cancers (Basel) 2017;9 doi: 10.3390/cancers9040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martina J.A., Diab H.I., Lishu I. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 2014;7 doi: 10.1126/scisignal.2004754. ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willett R., Martina J.A., Zewe J.P. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat. Commun. 2017;8:1580. doi: 10.1038/s41467-017-01871-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Y., Zou Z., Becker N. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugita S., Ito K., Yamashiro Y. EGFR-independent autophagy induction with gefitinib and enhancement of its cytotoxic effect by targeting autophagy with clarithromycin in non-small cell lung cancer cells. Biochem. Biophys. Res. Commun. 2015;461:28–34. doi: 10.1016/j.bbrc.2015.03.162. [DOI] [PubMed] [Google Scholar]
- 11.Ohtomo T., Miyazawa K., Naito M. Cytoprotective effect of imatinib mesylate in non-BCR-ABL-expressing cells along with autophagosome formation. Biochem. Biophys. Res. Commun. 2010;391:310–315. doi: 10.1016/j.bbrc.2009.11.055. https://doi:10.1016/j.bbrc.2009.11.055 [DOI] [PubMed] [Google Scholar]
- 12.Moriya S., Che X.F., Komatsu S. Macrolide antibiotics block autophagy flux and sensitize to bortezomib via endoplasmic reticulum stress-mediated CHOP induction in myeloma cells. Int. J. Oncol. 2013;42:1541–1550. doi: 10.3892/ijo.2013.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirasawa K., Moriya S., Miyahara K. Macrolide antibiotics exhibit cytotoxic effect under amino acid-depleted culture condition by blocking autophagy flux in head and neck squamous cell carcinoma cell lines. PloS One. 2016;11 doi: 10.1371/journal.pone.0164529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukai S., Moriya S., Hiramoto M. Macrolides sensitize EGFR-TKI-induced non-apoptotic cell death via blocking autophagy flux in pancreatic cancer cell lines. Int. J. Oncol. 2016;48:45–54. doi: 10.3892/ijo.2015.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaizuka T., Morishita H., Hama Y. An autophagic flux probe that releases an internal control. Mol. Cell. 2016;64:835–849. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 16.Saito Y., Moriya S., Kazama H. Amino acid starvation culture condition sensitizes EGFR-expressing cancer cell lines to gefitinib-mediated cytotoxicity by inducing atypical necroptosis. Int. J. Oncol. 2018;52:1165–1177. doi: 10.3892/ijo.2018.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Song Y., Quach C. Transcriptional regulation of autophagy-lysosomal function in BRAF-driven melanoma progression and chemoresistance. Nat. Commun. 2019;10:1693. doi: 10.1038/s41467-019-09634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galluzzi L., Vitale I., Aaronson S.A. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramakrishnan V., Timm M., Haug J.L. Sorafenib, a dual Raf kinase/vascular endothelial growth factor receptor inhibitor has significant anti-myeloma activity and synergizes with common anti-myeloma drugs. Oncogene. 2010;29:1190–1202. doi: 10.1038/onc.2009.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee C.S., Lee L.C., Yuan T.L. MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl. Acad. Sci. U.S.A. 2019 Feb 1 doi: 10.1073/pnas.1817494116. pii: 201817494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinsey C.G., Camolotto S.A., Boespflug A.M. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 2019;25:620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z., Chang P.C., Yang J.C. Autophagy blockade sensitizes prostate cancer cells towards Src family kinase inhibitors. Genes Cancer. 2010;1:40–49. doi: 10.1177/1947601909358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothschild S.I., Gautschi O., Batliner J. MicroRNA-106a targets autophagy and enhances sensitivity of lung cancer cells to Src inhibitors. Lung Canc. 2017;107:73–83. doi: 10.1016/j.lungcan.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Goodall M.L., Fitzwalter B.E., Zahedi S. The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev. Cell. 2016;37:337–349. doi: 10.1016/j.devcel.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galluzzi L., Kepp O., Kroemer G. MLKL regulates necrotic plasma membrane permeabilization. Cell Res. 2014;24:139–140. doi: 10.1038/cr.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang D., Zheng X., Wang Z.A. The MLKL channel in necroptosis is an octamer formed by tetramers in a dyadic process. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00497-16. pii:e00497-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.