Abstract
Ethanol-mediated down-regulation of carnitine palmitoyltransferase-1 (CPT-1A) gene expression plays a major role in the development of hepatic steatosis; however, the underlying mechanisms are not completely elucidated. Tributyrin, a butyrate prodrug that can inhibit histone deacetylase (HDAC) activity, attenuates hepatic steatosis and injury. The present study examined the beneficial effect of tributyrin/butyrate in attenuating ethanol-induced pathogenic epigenetic mechanisms affecting CPT-1A promoter–histone modifications and gene expression and hepatic steatosis/injury.
Methods
Mice were fed a liquid Lieber-DeCarli diet (Research Diet Inc, New Brunswick, NJ) with or without ethanol for 4 weeks. In a subset of mice, tributyrin (2 g/kg) was administered orally by gavage. Primary rat hepatocytes were treated with 50 mmol/L ethanol and/or 2 mmol/L butyrate. Gene expression and epigenetic modifications at the CPT-1A promoter were analyzed by chromatin immunoprecipitation analysis.
Results
In vivo, ethanol induced hepatic CPT-1A promoter histone H3K9 deacetylation, which is indicative of a repressive chromatin state, and decreased CPT-1A gene expression. Our data identified HDAC1 as the predominant HDAC causing CPT-1A promoter histone H3K9 deacetylation and epigenetic down-regulation of gene expression. Significantly, Specificity Protein 1 (SP1) and Hepatocyte Nuclear Factor 4 Alpha (HNF4α) participated in the recruitment of HDAC1 to the proximal and distal regions of CPT-1A promoter, respectively, and mediated transcriptional repression. Importantly, butyrate, a dietary HDAC inhibitor, attenuated ethanol-induced recruitment of HDAC1 and facilitated p300-HAT binding by enabling SP1/p300 interaction at the proximal region and HNF4α/peroxisomal proliferator-activated receptor-γ coactivator-1α/p300 interactions at the distal region, leading to promoter histone acetylation and enhanced CPT-1A transcription.
Conclusions
This study identifies HDAC1-mediated repressive epigenetic mechanisms that underlie an ethanol-mediated decrease in CPT-1A expression. Importantly, tributyrin/butyrate inhibits HDAC1, rescues CPT-1A expression, and attenuates ethanol-mediated hepatic steatosis and injury, suggesting its potential use in therapeutic strategies for alcoholic liver disease.
Keywords: Epigenetic Promoter Modification, Gene Regulation, Alcoholic Liver Disease, Hepatocytes
Abbreviations used in this paper: ALD, alcoholic liver disease; ChIP, chromatin immunoprecipitation; CPT-1A, carnitine palmitoyltransferase-1A; Ct, Cycle of threshold; DR, Direct Repeat; EF, ethanol-fed; HAT, histone acetyltransferase; HDAC, histone deacetylase; H3K9Ac, histone 3 lysine 9 acetylation; mRNA, messenger RNA; PCR, polymerase chain reaction; PGC-1α, Peroxisomal proliferator-activated receptor-γ coactivator-1α; Pol II, polymerase II; SP1, Specificity Protein 1; Tb, tributyrin; TRE, T3 response elements
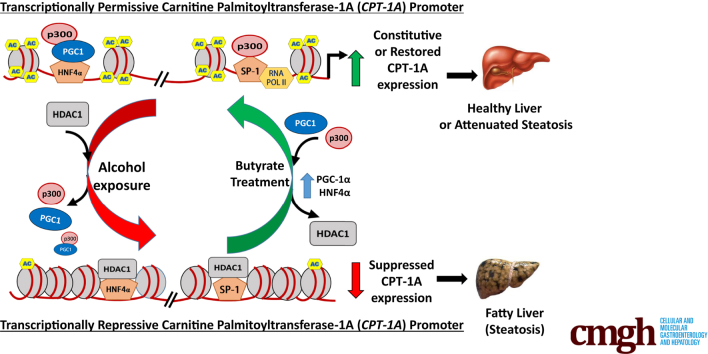
Graphical abstract
Summary.
Alcohol drinking inhibits carnitine palmitoyltransferase-1 expression by interfering with the interactions of key regulatory proteins on DNA via epigenetic mechanisms. These alcohol-mediated changes were prevented by oral administration of Tributyrin, a butyrate prodrug, thereby reducing fat accumulation and liver injury, indicating a therapeutic strategy in the treatment of alcoholic liver disease.
Alcoholic liver disease (ALD) is a multistage disease and is associated with alcohol-related mortality and morbidity.1, 2, 3 Hepatic steatosis is an early manifestation of ALD and has the potential to increase the susceptibility of the liver to secondary insults or injuries. Furthermore, alcohol-induced hepatic lipid accumulation can initiate and enhance the progression of ALD from steatosis to more severe forms of liver disease such as steatohepatitis and cirrhosis.1, 2, 3, 4 Although various pathways can contribute to hepatic steatosis, both experimental and clinical studies have reported that alcohol-induced impairment of β-oxidation plays a major role in the development of hepatic steatosis and liver disease.5 Hence, it is highly relevant to study the underlying molecular mechanisms involved in alcohol-induced dysregulation of β-oxidation leading to steatosis in the liver.
Hepatic mitochondrial β-oxidation is regulated mainly by carnitine palmitoyltransferase-1 (CPT-1A).6 CPT-1A acts as a rate-limiting enzyme and controls fatty acid transport into mitochondria for oxidation,7,8 and hence alterations in its expression/activity can affect hepatic fat accumulation. Studies have shown that alcohol feeding affects both function and expression of CPT-1. At a functional level, ethanol consumption decreases hepatic CPT-1 activity and also increases enzyme sensitivity to malonyl coenzyme A (malonyl-CoA) inhibition. Furthermore, this decrease in CPT-1 activity has been correlated with a decrease in the rate of fatty acid oxidation in hepatocytes, suggesting that CPT-1 may play a major role in the generation of ethanol-induced fatty liver.9, 10, 11 At the gene expression level, we and others have shown that alcohol consumption down-regulates CPT-1 expression, which plays a major role in the development of hepatic steatosis.12,13
There is gathering evidence regarding the contributory role of epigenetic mechanisms in the development of ALD.14,15 Epigenetic mechanisms, particularly promoter histone modifications, play a major role in controlling chromatin structure and regulate binding of transcription factors and transcriptional activation of genes. Although histone modifications, particularly acetylation, play a significant role in the development of alcohol-associated liver pathology, the underlying molecular epigenetic mechanisms affected by alcohol are only beginning to be understood.12,16,17 Promoter histone acetylation at the lysine residues regulated by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs) play an important role in maintaining the transcriptionally active and/or repressive chromatin state.18,19 Concerning CPT-1A expression, both proximal and distal regions of the CPT-1A promoter participate in the transcriptional regulation of CPT-1A gene expression.20, 21, 22, 23 Earlier work performed by us showed that acute alcohol-induced down-regulation of hepatic CPT-1A gene expression entails transcriptionally repressive promoter histone deacetylation, mediated by the Nuclear receptor co-repressor 1 (N-CoR)-HDAC3 corepressor complex, binding to the distal promoter region of the CPT-1A gene.12 However, in the context of chronic ethanol consumption, epigenetic mechanisms involving HDACs as well as HATs, affecting both proximal and distal promoter regulatory regions, leading to a decrease in CPT-1A gene expression, have not been determined.
There is increasing evidence that shows a significant pathogenic role for the gut-liver axis in the development of ALD.24 It has been documented that in the gut, alcohol decreases short chain fatty acids (particularly butyrate), which are produced mainly by microbial fermentation of indigestible dietary fibers.25 The decrease in short chain fatty acids likely occurs owing to alcohol-induced gut microbial dysbiosis.26 Studies have shown that butyrate has a protective role by acting as a HDAC inhibitor and affecting epigenetic regulation of gene expression.27 Tributyrin is a butyrate prodrug that after oral administration is hydrolyzed to butyrate and is able to increase plasma butyrate levels, providing HDAC inhibitory capabilities.28,29 Moreover, it has better pharmacokinetic properties with low toxicity than butyrate.30 Regarding beneficial effects, tributyrin (Tb) administration attenuates lipopolysaccharide and chronic binge alcohol-induced inflammatory reactions and gut-mediated liver injury.28,31 However, the epigenetic mechanisms underlying the hepatoprotective effects of Tb in ALD remain undetermined. Therefore, the present study examined the effect of oral Tb administration on HDAC and HAT promoter interactions, CPT-1A gene expression, and the development of hepatic steatosis and injury in an animal model of chronic ethanol feeding.
The data identify that Tb/butyrate, via their ability to inhibit HDAC function, prevents ethanol-induced repressive epigenetic mechanisms affecting CPT-1A promoter histone acetylation that down-regulates its expression and attenuates hepatic steatosis and injury. Importantly, the data imply that oral administration of Tb could be a potential component of the therapeutic strategy in the treatment of ALD.
Results
Tb Administration Attenuates Ethanol-Induced Hepatic Steatosis and Injury in Mice
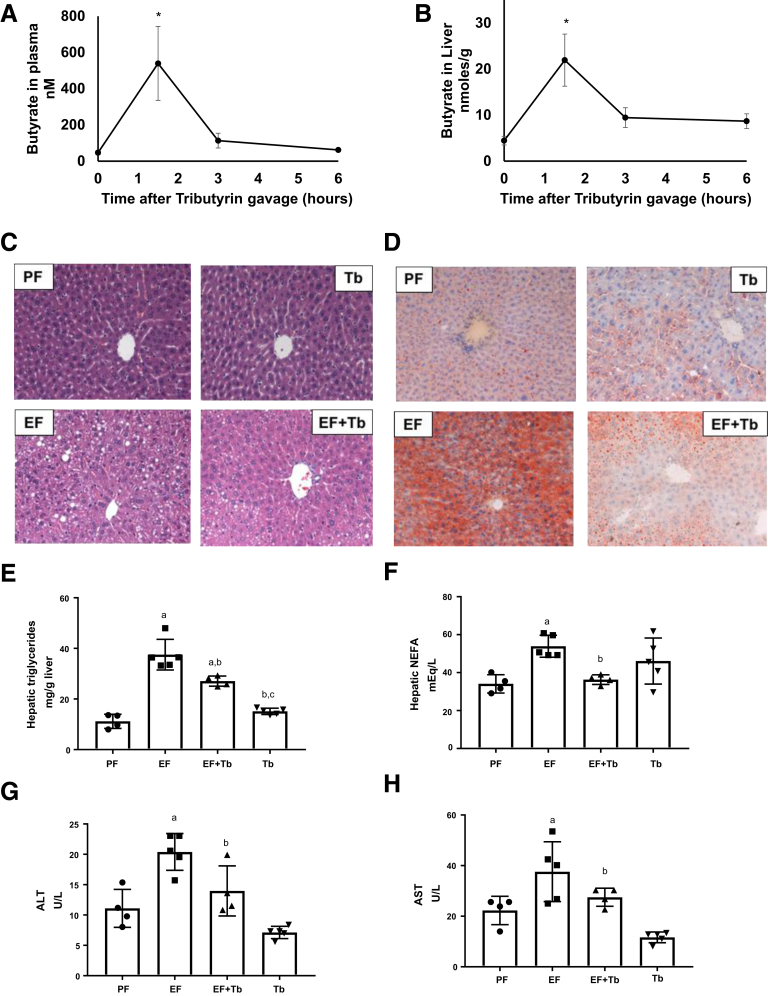
To investigate the efficacy of orally administrated Tb as a butyrate prodrug, we initially measured the butyrate levels in the portal blood and liver tissue in mice by gas chromatography–mass spectrometry. We used 2 g/kg of Tb, a concentration that protects against diet-induced hepatic steatosis.28 Pharmacokinetic analysis showed an increase in butyrate levels that peaked in the portal circulation (539.52 ± 204.05 nmol/L) and the liver tissue (21.93 ± 9.8 nmol/g) at 1.5 hours after oral administration. At 3 hours, serum butyrate levels rapidly decreased to baseline, while hepatic levels showed a trend of remaining slightly increased (Figure 1A and B). Furthermore, we examined the effect of Tb administration on ethanol-induced hepatic steatosis. As shown in Figure 1C and D, H&E and Oil Red O staining of liver sections showed that ethanol consumption resulted in considerable hepatic steatosis that was attenuated markedly in Tb-fed mice. Consistent with the observed decrease in lipid accumulation, reduced levels of hepatic triglycerides (Figure 1E) and free fatty acids (Figure 1F) also were detected in Tb-fed mice. Moreover, Tb significantly reduced an ethanol-mediated increase in serum alanine aminotransferase and aspartate aminotransferase levels (Figure 1G and H). These data indicate that oral administration of Tb significantly attenuates ethanol-induced hepatic steatosis and injury.
Figure 1.
Tb administration attenuates ethanol-induced hepatic steatosis and injury in mice. (A and B) Pharmacokinetic analysis of butyrate concentration in (A) plasma and (B) liver at 0, 1.5, 3, and 6 hours by gas chromatography with mass spectrometric detection in mice receiving oral gavage of Tb (2 g/kg). (C and D) Mice were fed a control diet (isocaloric maltose dextrin [PF]) or ethanol-containing (5% vol/vol) diet (EF) with or without administration of Tb by oral gavage (2 g/kg; Tb or EF + Tb) for 4 weeks. Representative micrographs show (C) H&E and (D) Oil Red O staining of liver sections (original magnification, 20×), showing fat accumulation in the liver of EF mice. Representative images are shown. Hepatic (E) triglycerides and (F) nonesterified fatty acids (NEFA) were assessed by biochemical analysis. Serum (G) alanine aminotransferase (ALT) and (H) aspartate aminotransferase (AST) levels were assessed for liver injury. Data were analyzed by analysis of variance and are represented as means ± SD, n = 4–6 animals/per group. P value < .05 for (a) when compared with PF, (b) when compared with EF, and (c) when compared with EF + Tb.
Tb Prevents an Ethanol-Induced Decrease in Hepatic CPT-1A Expression in Mice
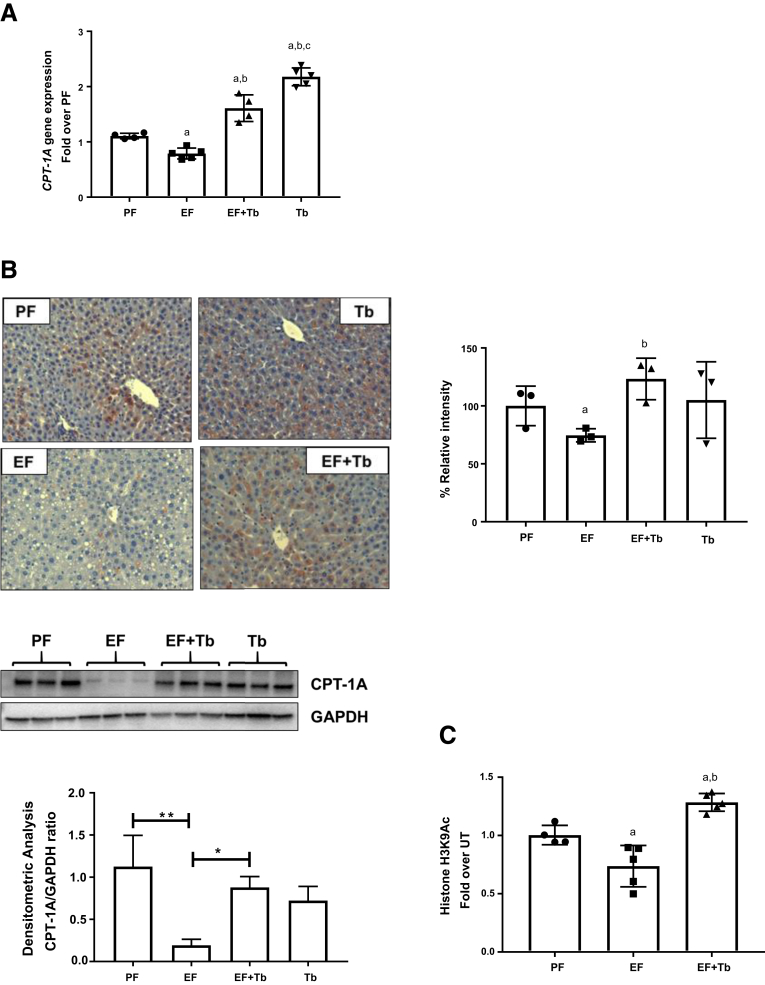
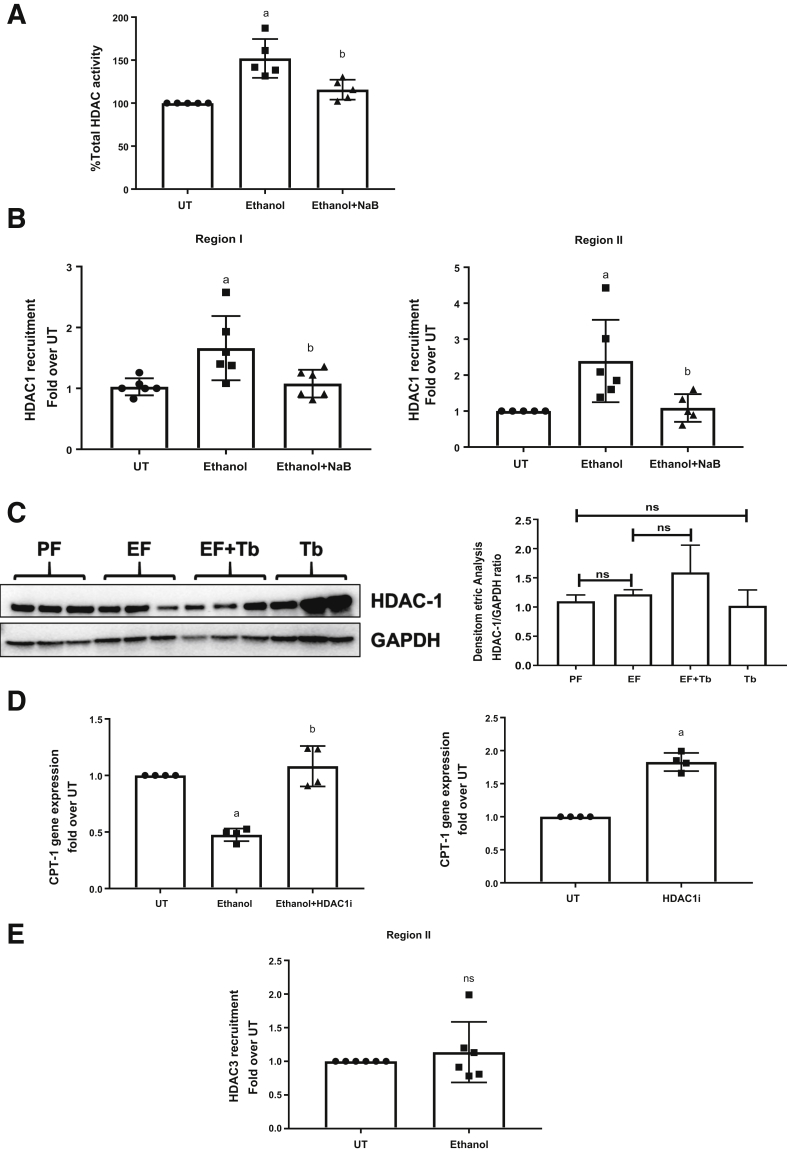
Work performed by us and others has shown that ethanol-mediated down-regulation of CPT-1A plays a major role in the development of hepatic steatosis.12,32, 33, 34 Hence, the effect of Tb on hepatic CPT-1A expression was assessed in ethanol-fed mice. In keeping with earlier reports, CPT-1A gene expression (messenger RNA [mRNA] and protein) was decreased significantly in ethanol-fed mice. Importantly, a reduction in CPT-1A expression in response to ethanol was significantly prevented by Tb administration (Figure 2A and 2B).
Figure 2.
Tb administration down-regulates hepatic CPT-1A expression in mice. Mice were pair-fed (isocaloric maltose dextrin [PF]) with either a control diet or EF (5% wt/vol) along with Tb (2 g/kg, EF + Tb) administered orally. (A) CPT-1A mRNA levels were analyzed by real-time quantitative PCR and normalized to β-actin mRNA. (B) Immunohistochemical staining with anti–CPT-1A antibody (original magnification, 20×). Representative images are shown. Bar graph shows the quantification of CPT-1A protein using the MetaMorph Microscopy Automation and Image Analysis Software (Molecular Devices, LLC, San Jose, CA) by calculating the percentage of positive (based on the average intensity of the field-of-view) microscope fields. Detection of CPT-1A protein levels by Western blot analysis in mice liver tissues. Blots were stripped and re-probed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody as a loading control. The density ratio for each band compared with its GAPDH is shown as a bar graph. Data are expressed as means ± SEM, n = 5 animals/per group. P value <.05 (a) when compared with PF, (b) when compared with EF. (C) ChIP analysis was performed on mice liver tissue. Chromatin fragments were immunoprecipitated with anti-H3K9Ac antibody and ChIP-qPCR was performed using primer pairs specific for the TRE region on the CPT-1A promoter. Nonimmunoprecipitated chromatin was used as input. Data were analyzed by analysis of variance and represented as means ± SD, n = 4–6 mice per group (n = 3 for Western blot). P value < .05 (a) when compared with PF, (b) when compared with EF.
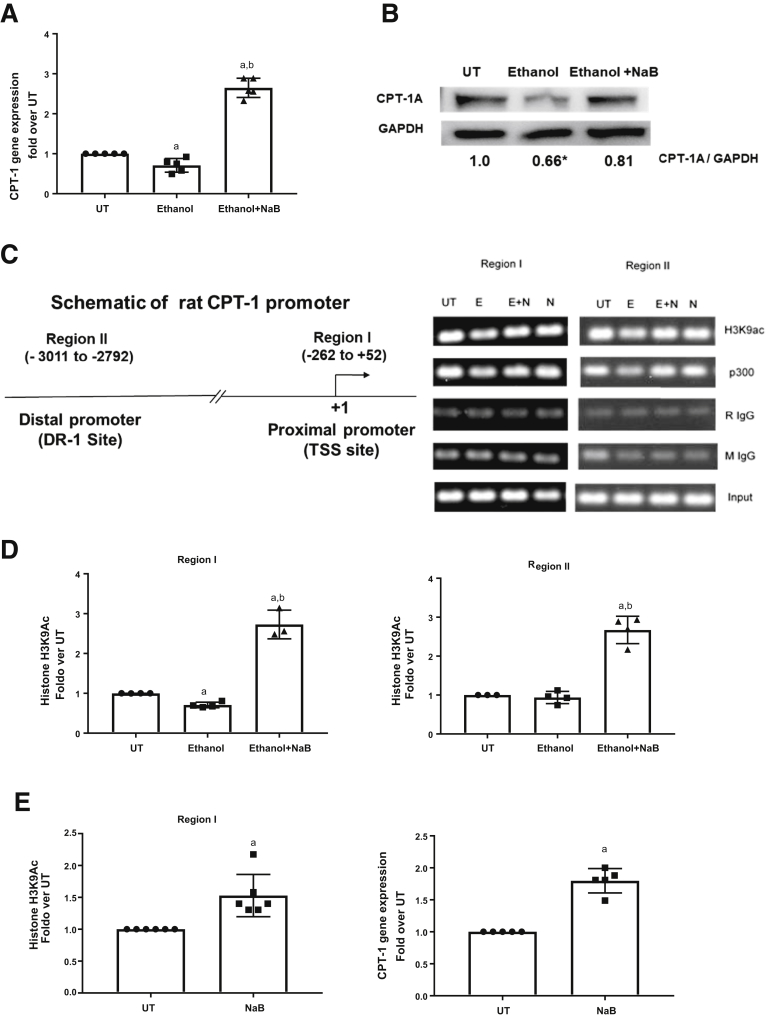
Our earlier work showed that binge ethanol exposure induced CPT-1A promoter histone H3 hypoacetylation, leading to a decrease in CPT-1A mRNA expression.12 Hence, we examined the effects of ethanol and butyrate on the status of histone H3 lysine 9 acetylation (H3K9Ac), which plays a key role in the transcriptional activation of gene expression. Examination of the CPT-1A proximal mouse promoter region (-169 to -81 bp) by chromatin immunoprecipitation (ChIP) analysis showed a significant decrease in histone H3K9Ac in ethanol-fed mice (EF). Significantly, Tb administration blocked this ethanol-induced H3K9 deacetylation (EF + Tb) (Figure 2C). These data suggest that Tb blocks transcriptionally repressive epigenetic mechanisms and prevents the decrease in CPT-1A mRNA expression.
Butyrate Inhibits Ethanol-Mediated HDAC1 Recruitment and Prevents the Decrease in CPT-1A Promoter Histone Deacetylation
To investigate the detailed epigenetic mechanisms that regulate hepatic promoter histone H3K9 acetylation and CPT-1A gene expression, we examined ethanol and butyrate inducible chromatin changes at the CPT-1A promoter in primary hepatocytes. Initially, CPT-1A gene expression was examined in hepatocytes either left untreated or treated with 50 mmol/L ethanol with or without sodium butyrate (2 mmol/L, 30 minutes before ethanol). Analogous to in vivo effects, butyrate prevented an ethanol-mediated decrease in CPT-1A gene expression at the mRNA as well as protein level (Figure 3A and B). ChIP analysis was studied in both proximal and distal promoter regions (region I: proximal transcription start site, -262 to +52; region II: distal DR-1 site, -3011 to -2792) of the CPT-1A gene (Figure 3C).6,12,23 Correspondent to gene expression, ChIP analysis showed that there was a modest yet significant reduction in transcriptionally permissive H3K9Ac by ethanol treatment at region I (Figure 3D). Butyrate pretreatment not only abrogated this decrease but substantively increased H3K9Ac by approximately 2.5-fold at regions I and II (Figure 3D). Moreover, butyrate treatment alone increased baseline H3K9Ac and CPT-1A mRNA expression in primary hepatocytes (Figure 3E).
Figure 3.
Butyrate prevents an ethanol-mediated decrease in CPT-1A gene expression and CPT-1A promoter–associated histone H3K9Ac. Primary hepatocytes were left untreated (UT) or treated with 50 mmol/L ethanol for 12 hours (Ethanol). Cells were pretreated with sodium butyrate (NaB 2 mmol/L) for 30 minutes before ethanol (Ethanol + NaB). (A) CPT-1A mRNA levels were analyzed by real-time quantitative PCR and normalized to β-actin mRNA. (B) Representative CPT-1A protein levels normalized to the corresponding control protein (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). Numbers represent the densitometry ratio normalized to GAPDH (n = 3). (C) Schematic representation of the location of key transcriptionally active regions (region I: proximal promoter-transcription start site [TSS], region II: distal promoter [DR-1] site) for analysis of epigenetic modifications are shown. The coordinate locations of regions were shown with respect to the +1 start site NCBI Reference Sequences (REFSEQ) NM_031559. (D) Chromatin fragments were immunoprecipitated with antibody specific for acetylated anti-H3K9Ac, and ChIP real-time quantitative PCR was performed using primer pairs specific for region I and II on the CPT-1A promoter. Nonimmunoprecipitated chromatin was used as input. Data were analyzed by analysis of variance and represented as means ± SD, n = 4–6 experiments. P value < .05 (a) when compared with UT, (b) when compared with ethanol. (E) ChIP assay and CPT-1A mRNA analysis was performed as detailed earlier only in sodium butyrate–treated primary hepatocytes.
To address the mechanisms underlying ethanol-mediated H3K9 deacetylation, the effects of ethanol and butyrate on HDAC activity were examined in primary hepatocytes (Figure 4A). Commensurate with its effect on H3K9Ac, ethanol treatment increased the total HDAC activity in primary hepatocytes. Importantly, butyrate, in accordance with its HDAC inhibitory function, prevented the ethanol-induced increase in HDAC activity. After an assessment of hepatocyte HDAC activity, we examined the effects of ethanol and butyrate on HDAC recruitment to the CPT-1A promoter.
Figure 4.
Butyrate prevents ethanol-mediated recruitment of HDAC1 to the CPT-1A promoter. Primary hepatocytes were left untreated (UT) or treated with 50 mmol/L ethanol for 12 hours (Ethanol). Cells were pretreated with sodium butyrate (NaB 2 mmol/L) for 30 minutes before ethanol (Ethanol + NaB). (A) HDAC activity was evaluated with the colorimetric HDAC activity assay kit. ChIP analysis was performed using chromatin fragments immunoprecipitated with (B) anti-HDAC1 and (E) anti-HDAC3. ChIP real-time quantitative PCR was performed using primer pairs specific for regions I and II on the CPT-1A promoter. Nonimmunoprecipitated chromatin was used as input. (C) Detection of HDAC-1 protein levels by Western blot analysis in mice liver tissues. Blots were stripped and reprobed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody as a loading control. The density ratio for each band compared with its GAPDH are shown in a bar graph. (D) H4IIE cells were kept UT or treated with 50 mmol/L ethanol for 24 hours. Cells were pretreated with HDAC1 inhibitor (HDAC1i) (1 μmol/L) 30 minutes before ethanol. CPT-1A mRNA levels were analyzed by reverse-transcription quantitative PCR and normalized to β-actin mRNA. Data were analyzed by analysis of variance and represented as means ± SD, n = 4 experiments. P value < .05 (a) when compared with UT, (b) when compared with ethanol, and (c) when compared with Ethanol + HDAC1i.
Transcription factors Specificity Protein 1 (SP1) and Hepatocyte Nuclear Factor 4 Alpha (HNF4α) are known to recruit HDAC1 to promoter regions and function as transcriptional repressors in a context-dependent manner.35,36 Because SP1 and HNF4α are established transcription factors that bind the CPT-1A promoter at regions I and II, respectively, we examined the effect of ethanol on HDAC1, SP1, and HNF4α recruitment in these regions. Consistent with decreased H3K9 acetylation and CPT-1A mRNA expression, ethanol treatment significantly increased HDAC1 binding at CPT-1A promoter regions I and II (Figure 4B). Importantly, butyrate abolished the ethanol-induced increase in HDAC1 binding to these regions (Figure 4B) and rescued H3K9 acetylation and CPT-1A mRNA expression. These changes in HDAC1 binding to the CPT-1A promoter regions was not accompanied by an increase or decrease in HDAC1 protein expression levels in any of the treatment conditions (Figure 4C). In addition, treatment of hepatocytes with a specific HDAC1 inhibitor prevented the ethanol-mediated down-regulation of CPT-1A mRNA expression, further supporting the suppressive role of HDAC1 (Figure 4D). Because HDAC3 has been shown by us and others to be recruited to the CPT-1A distal promoter region II via N-CoR binding,12,23 we also examined the effects of ethanol on HDAC3 recruitment at region II. The data showed that in comparison with HDAC1, there was no significant change in HDAC3 binding in ethanol-treated hepatocytes (Figure 4E).
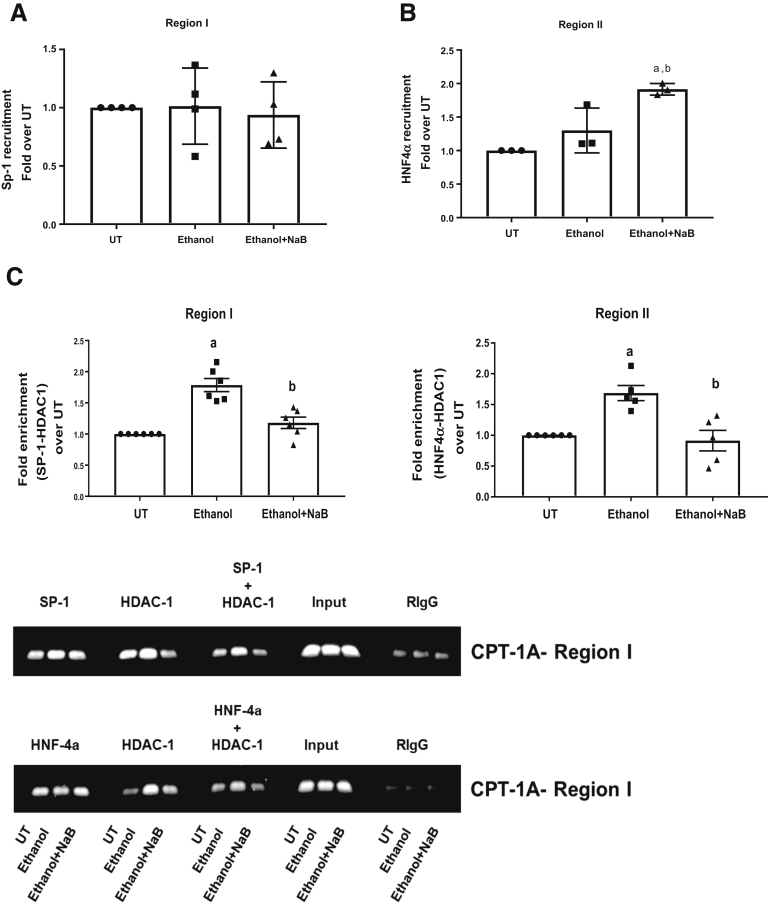
After the changes in HDAC1 binding to promoter regions I and II, we also examined the effects of ethanol and butyrate on the critical transcription factors SP1 and HNF4α that are known to bind the same promoter regions. Interestingly, ethanol treatment did not have any effect on either SP1 or HNF4α binding at region I or region II, respectively (Figure 5A and B), indicating a change in their ability to interact with HDAC1. Hence, we further examined the effect of ethanol on HDAC1 binding to SP1 and HNF4α by sequential ChIP/re-ChIP analysis. Specifically, chromatin harvested from treated primary hepatocytes was analyzed using either SP1 or HNF4α as the first ChIP antibody, followed by HDAC1 as the second (re-ChIP) antibody. The data showed that ethanol significantly increases the co-occupancy of HDAC1 with SP1 at region I and HNF4α at region II (Figure 5C). Importantly, pretreatment of butyrate significantly inhibited the interaction of HDAC1 to these transcription factors (Figure 5C). These data indicate that the ethanol exposure facilitated the interaction of SP1 and HNF4α with HDAC1, leading to its increased binding and transcriptionally repressive chromatin remodeling of the CPT-1A promoter.
Figure 5.
Ethanol does not affect recruitment of transcription factors but increases their interaction with HDAC1 to the CPT-1A promoter. Primary hepatocytes were left untreated (UT) or treated with 50 mmol/L ethanol for 12 hours (Ethanol). Cells were pretreated with sodium butyrate (NaB, 2 mmol/L) for 30 minutes before ethanol (Ethanol + NaB). ChIP analysis was performed using chromatin fragments immunoprecipitated with (A) anti-SP1 and (B) anti- HNF4α antibodies, and ChIP real-time quantitative PCR was performed using primer pairs specific for region I and region II, respectively, on the CPT-1A promoter. Nonimmunoprecipitated chromatin was used as input. Data were analyzed by analysis of variance and represented as means ± SD, n = 4. P value < .05 (a) when compared with UT and (b) when compared with ethanol. (C) Re-ChIP analysis shows the co-occupancy of transcription factors and HDAC-1 at the CPT-1A promoter. Chromatin was harvested from primary hepatocytes and re-ChIP was performed. The anti-SP1 or anti-HNF4α or Rabbit Immunoglobulin G (RIgG) antibodies were used for the first ChIP reaction followed by re-ChIP with anti-HDAC1 antibody. Quantitative ChIP-PCR analysis was performed in a similar manner as described earlier for ChIP analysis for both regions I and II of the CPT-1A promoter. Relative enrichment was normalized with input. The representative gel images are shown for each re-ChIP reaction. Data were analyzed by analysis of variance and represented as means ± SD, n = 4 experiments. P value < .05 (a) when compared with UT and (b) when compared with ethanol.
Butyrate Precludes Ethanol-Mediated Changes in Peroxisomal Proliferator-Activated Receptor-γ Coactivator-1α and p300 Recruitment to the CPT-1A Gene Promoter
Peroxisomal proliferator-activated receptor-γ coactivator-1α (PGC-1α) is a key transcriptional coactivator of CPT-1A gene expression that is affected by ethanol.37 PGC-1α is recruited to the distal direct repeat 1 (DR-1) region II of the CPT-1A promoter via its interaction with HNF4α and is known to enhance CPT-1A transcription.21 ChIP analysis showed that ethanol decreased PGC-1α binding significantly, which was prevented and further increased by butyrate treatment (Figure 6A). To address the butyrate-mediated enhancement of PGC-1α recruitment, we examined its effect on PGC-1α expression. Butyrate led to a marked increase in PGC-1α gene expression in ethanol-treated primary hepatocytes (Figure 6B and C). In addition, as shown earlier (Figure 5), butyrate treatment also increased the recruitment of HNF4α at the distal DR-1 region II site, potentially contributing to the enhanced PGC-1α binding (Figure 6A). Taken together, these data indicate that the ethanol-mediated decrease in PGC-1α expression and DNA binding led to a coordinated increase in HDAC1 recruitment to the CPT-1A promoter. Significantly, these ethanol effects are reversed by butyrate, via HDAC1 inhibition and increased PGC-1α expression and binding.
Figure 6.
Butyrate prevents an ethanol-mediated decrease in recruitment of PGC-1α and p300 to the CPT-1A promoter. Primary hepatocytes were left untreated (UT) or treated with 50 mmol/L ethanol for 12 hours (Ethanol). Cells were pretreated with sodium butyrate (NaB 2 mmol/L) for 30 minutes before ethanol (Ethanol + NaB). ChIP analysis was performed using chromatin fragments immunoprecipitated with (A) anti–PGC-1α and (D) anti-p300 antibodies. ChIP real-time quantitative PCR was performed using primer pairs specific for regions I and II on the CPT-1A promoter. Nonimmunoprecipitated chromatin was used as input. (C and E) Detection of PGC-1α and p300 protein levels by Western blot analysis in mice liver tissues. Blots were stripped and reprobed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody as a loading control. The density ratio for each band compared with its GAPDH are shown in an adjacent bar graph. (B) PGC-1α mRNA levels were analyzed by real time-qPCR and normalized to β-actin mRNA. Data were analyzed by analysis of variance and represented as means ± SD, n = 4 experiments. P value < .05 (a) when compared with UT and (b) when compared with ethanol.
PGC-1α as a co-activator can recruit HATs, particularly p300, which increases promoter histone acetylation and transcriptional activation.38 In addition, p300 also potentially can be recruited to the CPT-1A promoter region I via SP1 interaction.39 Hence, we examined p300 recruitment to the CPT-1A promoter at region II. In correlation with the decrease in PGC-1α binding, ethanol treatment also led to a decrease in p300 binding at region II (Figure 6D). Analogous to region II, ethanol treatment also significantly decreased p300 binding accompanied by a concurrent increase in HDAC1 binding at region I (Figure 6B). These data imply that the ethanol-mediated decrease in p300-HAT and a corresponding increase in HDAC1 binding culminate in CPT-1A promoter hypoacetylation. Significantly, butyrate treatment counters these HDAC1-mediated effects of ethanol by increasing p300 binding to both the distal and proximal regions of the CPT-1A promoter.
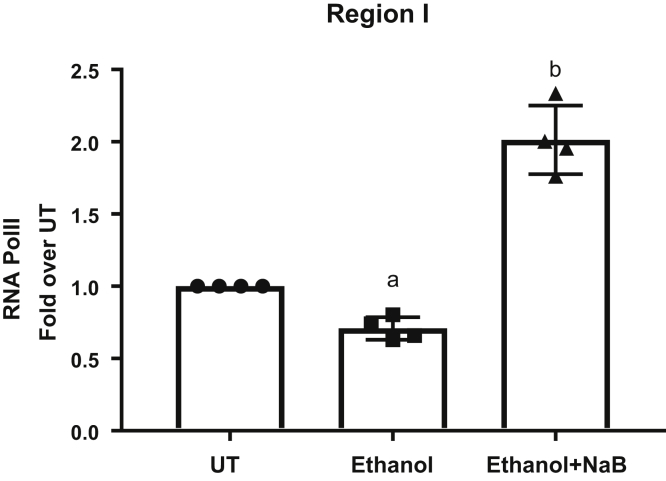
Butyrate Increases the Binding of RNA Polymerase II
In correlation with the decrease in PGC-1α and p300 binding and a simultaneous increase in HDAC1 binding and promoter histone hypoacetylation, ethanol disrupted the transcriptional complex as indicated by the decrease in the recruitment of RNA polymerase II (Pol II) at the CPT-1A promoter (Figure 7). Importantly, butyrate prevented these ethanol-induced repressive epigenetic effects and led to an increase in RNA Pol II recruitment and CPT-1A gene transcription (Figure 7).
Figure 7.
Butyrate increases RNA Pol II recruitment to the CPT-1A promoter. Primary hepatocytes were left untreated (UT) or treated with 50 mmol/L ethanol for 12 hours (Ethanol). Cells were pretreated with sodium butyrate (NaB, 2 mmol/L) for 30 minutes before ethanol (Ethanol + NaB). ChIP analysis was performed using chromatin fragments immunoprecipitated with anti-RNA Pol II antibody and ChIP real-time quantitative PCR was performed using primer pairs specific for region I on the CPT-1A promoter. Nonimmunoprecipitated chromatin was used as input. Data were analyzed by analysis of variance and represented as means ± SD, n = 4. P value < .05 (a) when compared with UT and (b) when compared with ethanol.
Discussion
Hepatic CPT-1A promotes the maintenance of lipid metabolism. Up-regulation in hepatic CPT-1A expression occurs in response to hormones and nutrients whereas down-regulation in hepatic CPT-1A expression is observed under alcohol or high-fat feeding conditions as well as altered circadian rhythms affecting cellular metabolism.12,40, 41, 42 However, the molecular mechanisms, specifically epigenetic chromatin modifications that contribute to ethanol-induced changes in CPT-1A transcription, have not been completely elucidated. In the present study, we identified CPT-1A promoter–associated epigenetic mechanisms that underlie both physiologic and ethanol-mediated pathologic deregulation of CPT-1A gene expression. Moreover, Tb protection against ethanol-mediated gut and liver damage has been reported, but its role as a dietary HDAC inhibitor on epigenetic regulation needs detailed investigation.43,44 Hence, we determined the therapeutic effects of butyrate that target ethanol-driven epigenetic deregulation of CPT-1A expression and attenuate hepatic steatosis.
We initially examined the status of the CPT-1A promoter histone acetylation, which indicates the transcriptionally permissive state of the chromatin (Figure 3). In accordance with the basal expression of CPT-1A, control untreated primary hepatocytes had demonstrable promoter histone H3K9Ac at both proximal and distal regions I and II. Significantly, under pathologic conditions of ethanol exposure, ethanol decreased promoter H3K9Ac levels and correspondingly decreased CPT-1A mRNA levels (Figure 3). These data suggest that ethanol induces promoter histone deacetylation, indicative of repressive chromatin configuration affecting CPT-1A mRNA expression. The status of promoter histone acetylation is regulated by the interplay between HATs and HDACs.18,19,45 Our data obtained identified that HDAC1 is the predominant HDAC recruited to the CPT-1A promoter, leading to histone deacetylation and down-regulation of gene expression. The protective effects of butyrate, a dietary HDAC inhibitor, further support the regulatory role of HDAC1. Butyrate inhibited the ethanol-induced recruitment of HDAC1 and consequent deacetylation and transcriptional suppression of CPT-1A. In addition, the effects of the HDAC1-specific inhibitor, which inhibited ethanol-induced down-regulation of CPT-1A mRNA expression, also supported the mechanistic role of HDAC1 (Figure 4).
Significantly, because HDACs do not directly bind DNA, the data obtained from our sequential ChIP/re-ChIP analysis showed that SP1 and HNF4α were involved in HDAC1 recruitment to the CPT-1A promoter at regions I and II in ethanol-treated hepatocytes (Figure 5). It is noteworthy that under normal physiological conditions both HNF4α and SP1 function as transcriptional factors that are required for hepatic CPT-1A mRNA expression.20,46 In view of this, our data show that in hepatocytes, ethanol alters the transcriptional function of SP1 and HNF4α, leading to the recruitment of HDAC1 to the CPT-1A promoter regions I and II, and mediate transcriptional repression. Indeed, both HNF4α and SP1, besides transcriptional activation, also have been shown to function as transcriptional repressors in a cell type, stimulus, and gene-specific manner.35,46,47 Taken together, the data indicate that in hepatocytes ethanol treatment can influence the function of HNF4α and SP1, leading to a transition from transcriptional activators to repressors that underlie ethanol-induced down-regulation of CPT-1A gene expression.
Concerning the protective effects, besides inhibiting HDAC1 binding, butyrate coordinately increased the binding of HNF4α along with the co-activator PGC-1α at the distal DR-1 region II (Figure 6). The effect of butyrate on increased HNF4α binding is significant because HNF4α is a major transcription factor that regulates CPT-1A transcription.21,46 Importantly, butyrate also increased the binding of PGC-1α, a transcriptional co-activator of CPT-1A that was decreased by ethanol. The interaction of HNF4α and PGC-1α at the distal DR-1 region II, which can occur in a ligand-independent manner, is important for the transcriptional activation of the CPT-1A gene.21,48 Of note, HNF4α transcriptional function is enhanced substantially when co-activated by PGC-1α.21,48 Butyrate, along with PGC-1α binding, also robustly activated PGC-1α mRNA expression in ethanol-treated hepatocytes. These findings indicate that the negative effects of ethanol on PGC-1α transcription and function also can be prevented by butyrate. Moreover, butyrate, similar to other short chain fatty acids, can act as a ligand for PPARα, activating its expression.49, 50, 51, 52 It also is known that PPAR can bind the intronic regions and regulate the inducible CPT-1A gene transcription and expression.53 Because ethanol was observed to decrease inducible PPARα expression and function,54, 55, 56 Tb/butyrate potentially also could counter this effect, thereby contributing to its protective function. The ethanol and butyrate-mediated epigenetic regulation of PGC-1α as well as PPARα gene expression and function currently is under investigation.
Although PGC-1α interacts and coactivates the nuclear-receptor transcription factor HNF4α, it lacks histone acetyltransferase activity required for histone acetylation and transcriptionally permissive chromatin function.38,57 Accordingly, in the context of CPT-1A gene expression, it has been suggested that PGC-1α further interacts with other co-activators.40 In particular, PGC-1α has been shown to interact with HAT coactivators including p300, and function cooperatively to mediate promoter histone acetylation and transcriptional activation of certain genes.38,57 In this regard, the data obtained showed that an ethanol-induced decrease in PGC-1α binding also led to a concomitant decrease in the recruitment of p300-HAT and a consequent decrease in basal H3K9 acetylation and CPT-1A transcription. Besides the distal DR-1 region II, p300-HAT also potentially could be recruited to the proximal promoter region I via its interaction with the transcription factor SP1. p300-HAT can act as a co-activator for SP1-mediated transcription effecting promoter histone acetylation.58 Concerning CPT-1A transcription, SP1 binds the proximal region (approximately -200 nucleotides) of the TATA-less CPT-1A gene and plays an essential role in its basal expression.20 The findings obtained show that p300-HAT interacts with SP1 in region I and participates in promoter histone acetylation and basal transcription of CPT-1A.
Similar to the inhibitory effect on p300-HAT recruitment at region II, ethanol also decreased its binding to region I, which correlated with a decrease in promoter H3K9 acetylation and CPT-1A transcription. Significantly, butyrate clearly blocked the negative effects of ethanol, enabling HNF4α/PGC-1α/p300 interactions at the distal DR-1 region II and SP1/p300 interaction at proximal region I, leading to promoter histone acetylation and CPT-1A transcription.
In conclusion, the present studies provide evidence for the ethanol-induced, HDAC1-mediated, transcriptionally repressive, epigenetic mechanisms that down-regulate CPT-1A gene expression contributing to hepatic steatosis. Our results also show that Tb/butyrate effectively can inhibit ethanol-induced HDAC1 recruitment to CPT-1A promoter regions and suppression of gene expression, attenuating hepatic steatosis and injury. Moreover, examination of the chromatin state of the CPT-1A promoter region in primary hepatocytes under basal as well as ethanol and butyrate treatment conditions also identified the interplay of major molecular components, namely HNF4α, PGC-1α, SP1, and p300-HAT in regulating CPT-1A transcription. Taken together, these data identify the pathogenic role of HDAC1 in ethanol-induced hepatic steatosis and injury that may serve as a potential therapeutic target in the treatment strategy for ALD.
Materials and Methods
Animal Model
Eight-week-old male C57BL/6N mice were obtained from Harlan (Indianapolis, IN). All mice were housed in a pathogen-free, temperature-controlled animal facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care with 12-hour light–dark cycles. All experiments were performed according to the criteria outlined in the Guide for Care and Use of Laboratory Animals and with the approval of the University of Louisville Animal Care and Use Committee. Mice were fed a modified Lieber-DeCarli liquid diet enriched in unsaturated fat (corn oil), which provided 40% of energy from fat, 43% from carbohydrates, and 17% from protein (Research Diet, New Brunswick, NJ).
Mice were pair-fed a Lieber-DeCarli liquid diet containing either ethanol or isocaloric maltose dextrin for 4 weeks. Ethanol was increased gradually over a period of 1 week and then mice were fed the ethanol diet (5% vol/vol) ad libitum for 4 weeks. The control pair-fed mice were given the isocaloric maltose-dextrin–containing liquid diet. For Tb treatment groups, both pair-fed (Tb) as well as ethanol-fed (EF + Tb) mice received Tb (Sigma Aldrich, St. Louis, MO) by oral gavage (2 g/kg, 5 days/wk) for 4 weeks.
Analysis of Butyrate by Gas Chromatography–Mass Spectrometry
Plasma samples were derivatized with pentafluorobenzyl bromide and extracted with hexane. The hexane extract was analyzed by gas chromatography–mass spectrometry using an Agilent DB-225 J&W GC column (Agilent Technologies Inc, Santa Clara, CA) with a temperature gradient from 50°C to 220°C at 30°C/min, at a flow rate of 1.5 mL/min with helium as a carrier gas. Detection was by electron ionization and selective ion monitoring of m/z at 256 for sample and m/z at 257 for 13C-labeled butyrate standard. Using the peak area for corresponding labeled and unlabeled derivatives, a standard curve was generated by plotting 12C/13C ratios against the concentration of solutions. The slope and intercept were used to determine the concentration of butyrate in the samples.
Liver Histopathologic Examination
For histologic analysis, liver sections were fixed in 10% buffered formalin for 24 hours and embedded in paraffin. Tissue sections were stained with H&E and examined under light microscopy.
Oil Red O Staining
To examine the amount of fat accumulation, the liver sections were stained with Oil Red O. Frozen liver sections were washed in phosphate-buffered saline twice for 5 minutes. Oil Red O and 85% propylene glycol were added with agitation for 15 minutes, followed by washing in tap water.
Biochemical Analyses
Total liver triglycerides were extracted from mouse liver tissue and quantified using triglyceride reagents (Thermo Fisher Scientific, Inc, Waltham, MA) as previously described.12 Briefly, hepatic tissue (100 mg) was homogenized in 50 mmol/L NaCl. The homogenate (500 μL) was mixed with chloroform/methanol (2:1, 4 mL) and incubated overnight at room temperature with gentle shaking. Homogenates were vortexed and centrifuged for 5 minutes at 3000 Χ g. The lower lipid phase was collected and concentrated by vacuum. The lipid pellets were dissolved in 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO) in phosphate-buffered saline, and hepatic triglyceride content was determined via enzymatic colorimetric methods.
Immunohistochemical Staining
Mice liver sections were stained with a commercially available antibody against CPT-1A (Proteintech Group, Inc, Chicago, IL) according to the manufacturer’s protocols.
Cell Culture and Treatments
All cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 10 U/mL penicillin, and 10 g/mL streptomycin. Cells were maintained in an incubator at 37°C with 5% CO2. Cells were plated and incubated overnight before treatment, in all experiments. Cells were untreated and/or treated with ethanol (50 mmol/L) for the indicated times. Sodium butyrate (2 mmol/L) treatment was performed 30 minutes before ethanol exposure (ethanol + sodium butyrate). The HDAC inhibitor entinostat was purchased from Selleckchem (Houston, TX).
RNA Isolation and Real-Time Polymerase Chain Reaction Analysis
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA) and complementary DNA was made using Quanta qScript (Quanta BioSciences, Gaithersburg, MD). The real-time polymerase chain reaction (PCR) was performed with Quanta Perfecta SYBR green fast mix and the ABI prism 7500 sequence detection system (Applied Biosystems, Foster City, CA). The relative gene expression was analyzed using the delta delta cycle of threshold 2-ΔΔCt method by normalizing with β-actin gene expression in all the experiments and is presented as fold change over untreated/pair-fed treatment group, which was set at 1.
Rat CPT-1A mRNA primers were as follows: forward: CTGCATGGAAGATGCTTTGA, reverse: GCCATGACATACTCCCACAA; mouse CPT-1A mRNA primers were as follows: forward: GCTGCACTCCTGGAAGAAGA, reverse: GGAGGGGTCCACTTTGGTAT; and mouse/rat β-actin mRNA primers were as follows: forward: CAGCTGAGAGGGAAATCGTG, reverse: CTCCAGGGAGGAAGAGGATG.
Total HDAC Activity
Total HDAC activity in the primary hepatocytes was estimated using a commercially available HDAC activity/inhibition assay kit (colorimetric) according to the manufacturer's protocol (Epigentek, Farmingdale, NY).
ChIP and Quantitative ChIP PCR Analysis
The ChIP assay was conducted using the ChIP assay protocol established in the laboratory.59 ChIP antibodies directed against anti-acetyl H3K9 (17-615), anti-HDAC1(17-608), anti-HDAC3 (17-10238), anti-p300 (05-257), anti-SP1 (17-601), and anti-RNA polymerase II (17-672) were purchased from EMD Millipore (Burlington, MA), anti–PGC-1α (Sc-13067) (Santa Cruz, Dallas, TX), and anti-HNF4α (ab41898) (Abcam, Cambridge, United Kingdom). ChIP-PCR primers designed for the regions of the CPT-1A were used and their sequences are detailed later. Data were analyzed as the differential occupancy fold change. ChIP–quantitative real-time PCR results were calculated by the ΔΔCt method in which each ChIP DNA fraction’s Ct value was normalized to the input DNA fraction. Semiquantitative ChIP PCR was performed and analyzed by ethidium bromide–stained agarose gel electrophoresis using CPT-1A promoter–specific primers (Figure 3C).
Three ChIP-PCR primers were designed for each region of the rat CPT-1 promoter and 1 primer for the mouse CPT-1 promoter. The primer sequences were as follows: region I of the mouse CPT-1 promoter: TRE forward: GGTGACGTTGGCTGAGCAA and TRE reverse: TGAGCCCCTGTACACGTTTTG; region I of the rat CPT-1 promoter: Chp2_forward: ATGGGCATGGCTTTAATGAG and Chp2_reverse: GGCTAGGACCCGAGCTTGT; GCTSspn_forward: AGCCTCGCCCGCCCCTGCTC and GCTSspn_reverse: CAGCGCTGCCCTCCCGGTGTC; and GCTSspn_forward: TCCAGGCCCCGCCCCGTCCT and GCTSspn_reverse: GCGCCGCGGGTGATTGGCTGA. Region II of the rat CPT-1 promoter: TRE forward: GGTGACGTTGGCTGAGCAA and TRE reverse: TGAGCCCCTGTACACGTTTTG; TrChp_forward: GCCTCATGGACTCCAAGTTC and TrChp_reverse: TATTTGTTCAGCCAGCGTCA; and TrChp_forward: CAAAACGTGTACAGGAGCTCAA and CPT-1 TrChp_reverse: TAATCCCAGAAGGCAGTGCT.
Sequential ChIP/Re-ChIP Analysis
The re-ChIP method was performed as described previously60 to detect HDAC-1 binding with transcription factors. Briefly, the chromatin was prepared and the first ChIP reaction was set up for anti–SP-1, anti-HNF4α, and anti-IgG control as mentioned in the ChIP assay method earlier. After the overnight incubation, the beads were pelleted and washed once each with buffer 1 (0.1% sodium dodecyl sulfate, 1% Triton X-100, 2 mmol/L EDTA, 20 mmol/L Tris–HCl [pH 8.0], 150 mmol/L NaCl), buffer 2 (0.1% sodium dodecyl sulfate, 1% Triton X-100, 2 mmol/L EDTA, 20 mmol/L Tris–HCl [pH 8.0], 500 mmol/L NaCl), buffer 3 (1% NP40, 1% deoxycholic acid, 1 mmol/L EDTA, 10 mmol/L Tris–HCl [pH 8.0], 250 mmol/L LiCl), and buffer 4 (2 mmol/L EDTA, 10 mmol/L Tris–HCl [pH 8.0]). After the final wash, the DNA–protein complexes were eluted with 50 μL 10 mmol/L dithiothreitol at 37°C for 30 minutes with gentle shaking. The beads were pelleted, the supernatant was diluted at least 20 times with ChIP dilution buffer, and the secondary ChIP reaction was performed using anti–HDAC-1 antibody. After overnight incubation with the second antibody, the wash steps with buffers 1–4 were repeated. The DNA was eluted from the beads and purified similar to the ChIP assay. The ChIP- PCR primers designed for regions I and II of the CPT-1A promoter were used to determine the co-occupancy of the transcription factor and HDAC-1. The fold enrichment of co-occupied protein was detected using the ΔΔCt method in which each ChIP DNA fraction’s Ct value was normalized to the input DNA fraction as described in the ChIP and quantitative ChIP PCR analysis method.
Statistical Analysis
Data are presented as means ± SEM for the indicated number of independently performed experiments or 4–6 mice per group. The Student t test and 1-way analysis of variance with Bonferroni multiple comparison tests were used for the determination of statistical significance. P < .05 was considered significant.
All authors had access to all data and have reviewed and approved the final manuscript.
Acknowledgements
The authors thank Dr Laura Nagy and Ms Marion McClain for a careful review of the manuscript and providing excellent suggestions.
Footnotes
Author contributions Shirish Barve was responsible for the study design and concept; Hridgandh Donde and Smita Ghare performed experiments; JingWen Zhang performed the animal histology analysis; Manicka V. Vadhanam, Pawel Lorkiewicz, and Sanjay Srivastava performed the gas chromatography–mass spectrometry analysis; Smita Ghare, Hridgandh Donde, Leila Gobejishvili, Craig J. McClain, and Shirish Barve analyzed and interpreted data; Smita Ghare, Hridgandh Donde, Swati Joshi-Barve, Craig J. McClain, and Shirish Barve prepared the manuscript; and all authors discussed the results and critically commented on the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health awards P20GM113226 (C.J.M. and S.B.) and P20GM103492 (S.S.); National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health awards P50AA024337 (C.J.M. and S.B.), RO1AA024405 (S.B.), and UO1AA022618 (S.B.); and National Institute of Environmental Health Sciences award P42ES023716 (S.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Dugum M., McCullough A. Diagnosis and management of alcoholic liver disease. J Clin Transl Hepatol. 2015;3:109–116. doi: 10.14218/JCTH.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathurin P., Bataller R. Trends in the management and burden of alcoholic liver disease. J Hepatol. 2015;62(Suppl):S38–S46. doi: 10.1016/j.jhep.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singal A.K., Bataller R., Ahn J., Kamath P.S., Shah V.H. ACG clinical guideline: alcoholic liver disease. Am J Gastroenterol. 2018;113:175–194. doi: 10.1038/ajg.2017.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day C.P., James O.F. Hepatic steatosis: innocent bystander or guilty party? Hepatology. 1998;27:1463–1466. doi: 10.1002/hep.510270601. [DOI] [PubMed] [Google Scholar]
- 5.Donohue T.M., Jr. Alcohol-induced steatosis in liver cells. World J Gastroenterol. 2007;13:4974–4978. doi: 10.3748/wjg.v13.i37.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook G.A., Edwards T.L., Jansen M.S., Bahouth S.W., Wilcox H.G., Park E.A. Differential regulation of carnitine palmitoyltransferase-I gene isoforms (CPT-I alpha and CPT-I beta) in the rat heart. J Mol Cell Cardiol. 2001;33:317–329. doi: 10.1006/jmcc.2000.1304. [DOI] [PubMed] [Google Scholar]
- 7.McGarry J.D., Brown N.F. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 8.Kerner J., Hoppel C. Fatty acid import into mitochondria. Biochim Biophys Acta. 2000;1486:1–17. doi: 10.1016/s1388-1981(00)00044-5. [DOI] [PubMed] [Google Scholar]
- 9.Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto T. Peroxisomal beta-oxidation enzymes. Neurochem Res. 1999;24:551–563. doi: 10.1023/a:1022540030918. [DOI] [PubMed] [Google Scholar]
- 11.Guzman M., Geelen M.J. Effects of ethanol feeding on the activity and regulation of hepatic carnitine palmitoyltransferase I. Arch Biochem Biophys. 1988;267:580–588. doi: 10.1016/0003-9861(88)90065-3. [DOI] [PubMed] [Google Scholar]
- 12.Kirpich I., Zhang J., Gobejishvili L., Kharebava G., Barker D., Ghare S., Joshi-Barve S., McClain C.J., Barve S. Binge ethanol-induced HDAC3 down-regulates Cpt1alpha expression leading to hepatic steatosis and injury. Alcohol Clin Exp Res. 2013;37:1920–1929. doi: 10.1111/acer.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li K., Xiao Y., Yu J., Xia T., Liu B., Guo Y., Deng J., Chen S., Wang C., Guo F. Liver-specific gene inactivation of the transcription factor ATF4 alleviates alcoholic liver steatosis in mice. J Biol Chem. 2016;291:18536–18546. doi: 10.1074/jbc.M116.726836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugimoto K., Takei Y. Pathogenesis of alcoholic liver disease. Hepatol Res. 2017;47:70–79. doi: 10.1111/hepr.12736. [DOI] [PubMed] [Google Scholar]
- 15.Moghe A., Joshi-Barve S., Ghare S., Gobejishvili L., Kirpich I., McClain C.J., Barve S. Histone modifications and alcohol-induced liver disease: are altered nutrients the missing link? World J Gastroenterol. 2011;17:2465–2472. doi: 10.3748/wjg.v17.i20.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park P.H., Lim R.W., Shukla S.D. Gene-selective histone H3 acetylation in the absence of increase in global histone acetylation in liver of rats chronically fed alcohol. Alcohol Alcohol. 2012;47:233–239. doi: 10.1093/alcalc/ags004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kendrick S.F., O'Boyle G., Mann J., Zeybel M., Palmer J., Jones D.E., Day C.P. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology. 2010;51:1988–1997. doi: 10.1002/hep.23572. [DOI] [PubMed] [Google Scholar]
- 18.Peserico A., Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberharter A., Becker P.B. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steffen M.L., Harrison W.R., Elder F.F., Cook G.A., Park E.A. Expression of the rat liver carnitine palmitoyltransferase I (CPT-Ialpha) gene is regulated by Sp1 and nuclear factor Y: chromosomal localization and promoter characterization. Biochem J. 1999;340:425–432. [PMC free article] [PubMed] [Google Scholar]
- 21.Louet J.F., Hayhurst G., Gonzalez F.J., Girard J., Decaux J.F. The coactivator PGC-1 is involved in the regulation of the liver carnitine palmitoyltransferase I gene expression by cAMP in combination with HNF4 alpha and cAMP-response element-binding protein (CREB) J Biol Chem. 2002;277:37991–38000. doi: 10.1074/jbc.M205087200. [DOI] [PubMed] [Google Scholar]
- 22.Wei T., Xiong F.F., Wang S.D., Wang K., Zhang Y.Y., Zhang Q.H. Flavonoid ingredients of Ginkgo biloba leaf extract regulate lipid metabolism through Sp1-mediated carnitine palmitoyltranferase 1A up-regulation. J Biomed Sci. 2014;21:87. doi: 10.1186/s12929-014-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alenghat T., Meyers K., Mullican S.E., Leitner K., Adeniji-Adele A., Avila J., Bucan M., Ahima R.S., Kaestner K.H., Lazar M.A. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabo G. Gut-liver axis in alcoholic liver disease. Gastroenterology. 2015;148:30–36. doi: 10.1053/j.gastro.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie G., Zhong W., Zheng X., Li Q., Qiu Y., Li H., Chen H., Zhou Z., Jia W. Chronic ethanol consumption alters mammalian gastrointestinal content metabolites. J Proteome Res. 2013;12:3297–3306. doi: 10.1021/pr400362z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bull-Otterson L., Feng W., Kirpich I., Wang Y., Qin X., Liu Y., Gobejishvili L., Joshi-Barve S., Ayvaz T., Petrosino J., Kong M., Barker D., McClain C., Barve S. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davie J.R. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(Suppl):2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi M., Sakaki H., Usami M., Iizuka N., Shuno K., Aoyama M., Usami Y. Oral administration of tributyrin increases concentration of butyrate in the portal vein and prevents lipopolysaccharide-induced liver injury in rats. Clin Nutr. 2011;30:252–258. doi: 10.1016/j.clnu.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 29.Heidor R., Furtado K.S., Ortega J.F., de Oliveira T.F., Tavares P.E., Vieira A., Miranda M.L., Purgatto E., Moreno F.S. The chemopreventive activity of the histone deacetylase inhibitor tributyrin in colon carcinogenesis involves the induction of apoptosis and reduction of DNA damage. Toxicol Appl Pharmacol. 2014;276:129–135. doi: 10.1016/j.taap.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Newmark H.L., Lupton J.R., Young C.W. Butyrate as a differentiating agent: pharmacokinetics, analogues and current status. Cancer Lett. 1994;78:1–5. doi: 10.1016/0304-3835(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 31.Cresci G.A., Glueck B., McMullen M.R., Xin W., Allende D., Nagy L.E. Prophylactic tributyrin treatment mitigates chronic-binge alcohol-induced intestinal barrier and liver injury. J Gastroenterol Hepatol. 2017;32:1587–1597. doi: 10.1111/jgh.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ajmo J.M., Liang X., Rogers C.Q., Pennock B., You M. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G833–G842. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong W.I., Osei-Hyiaman D., Park O., Liu J., Batkai S., Mukhopadhyay P., Horiguchi N., Harvey-White J., Marsicano G., Lutz B., Gao B., Kunos G. Paracrine activation of hepatic CB1 receptors by stellate cell-derived endocannabinoids mediates alcoholic fatty liver. Cell Metab. 2008;7:227–235. doi: 10.1016/j.cmet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Kirpich I., Ghare S., Zhang J., Gobejishvili L., Kharebava G., Barve S.J., Barker D., Moghe A., McClain C.J., Barve S. Binge alcohol-induced microvesicular liver steatosis and injury are associated with down-regulation of hepatic Hdac 1, 7, 9, 10, 11 and up-regulation of Hdac 3. Alcohol Clin Exp Res. 2012;36:1578–1586. doi: 10.1111/j.1530-0277.2012.01751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doetzlhofer A., Rotheneder H., Lagger G., Koranda M., Kurtev V., Brosch G., Wintersberger E., Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol. 1999;19:5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanulovic V.S., Kyrmizi I., Kruithof-de Julio M., Hoogenkamp M., Vermeulen J.L., Ruijter J.M., Talianidis I., Hakvoort T.B., Lamers W.H. Hepatic HNF4alpha deficiency induces periportal expression of glutamine synthetase and other pericentral enzymes. Hepatology. 2007;45:433–444. doi: 10.1002/hep.21456. [DOI] [PubMed] [Google Scholar]
- 37.Lieber C.S., Leo M.A., Wang X., Decarli L.M. Effect of chronic alcohol consumption on hepatic SIRT1 and PGC-1alpha in rats. Biochem Biophys Res Commun. 2008;370:44–48. doi: 10.1016/j.bbrc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Wallberg A.E., Yamamura S., Malik S., Spiegelman B.M., Roeder R.G. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 39.Hung J.J., Wang Y.T., Chang W.C. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol Cell Biol. 2006;26:1770–1785. doi: 10.1128/MCB.26.5.1770-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Ma K., Song S., Elam M.B., Cook G.A., Park E.A. Peroxisomal proliferator-activated receptor-gamma coactivator-1 alpha (PGC-1 alpha) enhances the thyroid hormone induction of carnitine palmitoyltransferase I (CPT-I alpha) J Biol Chem. 2004;279:53963–53971. doi: 10.1074/jbc.M406028200. [DOI] [PubMed] [Google Scholar]
- 41.Vancura P., Wolloscheck T., Baba K., Tosini G., Iuvone P.M., Spessert R. Circadian and dopaminergic regulation of fatty acid oxidation pathway genes in retina and photoreceptor cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyman L.R., Tian L., Hamm D.A., Schoeb T.R., Gower B.A., Nagy T.R., Wood P.A. Long term effects of high fat or high carbohydrate diets on glucose tolerance in mice with heterozygous carnitine palmitoyltransferase-1a (CPT-1a) deficiency: diet influences on CPT1a deficient mice. Nutr Diabetes. 2011;1:e14. doi: 10.1038/nutd.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cresci G.A., Glueck B., McMullen M.R., Xin W., Allende D., Nagy L.E. Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J Gastroenterol Hepatol. 2017;32:1587–1597. doi: 10.1111/jgh.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cresci G.A., Bush K., Nagy L.E. Tributyrin supplementation protects mice from acute ethanol-induced gut injury. Alcohol Clin Exp Res. 2014;38:1489–1501. doi: 10.1111/acer.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legube G., Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4:944–947. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Jimenez C.P., Kyrmizi I., Cardot P., Gonzalez F.J., Talianidis I. Hepatocyte nuclear factor 4alpha coordinates a transcription factor network regulating hepatic fatty acid metabolism. Mol Cell Biol. 2010;30:565–577. doi: 10.1128/MCB.00927-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar P., Tripathi S., Pandey K.N. Histone deacetylase inhibitors modulate the transcriptional regulation of guanylyl cyclase/natriuretic peptide receptor-a gene: interactive roles of modified histones, histone acetyltransferase, p300, AND Sp1. J Biol Chem. 2014;289:6991–7002. doi: 10.1074/jbc.M113.511444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhee J., Inoue Y., Yoon J.C., Puigserver P., Fan M., Gonzalez F.J., Spiegelman B.M. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higashimura Y., Naito Y., Takagi T., Uchiyama K., Mizushima K., Yoshikawa T. Propionate promotes fatty acid oxidation through the up-regulation of peroxisome proliferator-activated receptor alpha in intestinal epithelial cells. J Nutr Sci Vitaminol (Tokyo) 2015;61:511–515. doi: 10.3177/jnsv.61.511. [DOI] [PubMed] [Google Scholar]
- 50.Weng H., Endo K., Li J., Kito N., Iwai N. Induction of peroxisomes by butyrate-producing probiotics. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russo R., De Caro C., Avagliano C., Cristiano C., La Rana G., Mattace Raso G., Berni Canani R., Meli R., Calignano A. Sodium butyrate and its synthetic amide derivative modulate nociceptive behaviors in mice. Pharmacol Res. 2016;103:279–291. doi: 10.1016/j.phrs.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 52.Alex S., Lange K., Amolo T., Grinstead J.S., Haakonsson A.K., Szalowska E., Koppen A., Mudde K., Haenen D., Al-Lahham S., Roelofsen H., Houtman R., van der Burg B., Mandrup S., Bonvin A.M., Kalkhoven E., Muller M., Hooiveld G.J., Kersten S. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2013;33:1303–1316. doi: 10.1128/MCB.00858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song S., Attia R.R., Connaughton S., Niesen M.I., Ness G.C., Elam M.B., Hori R.T., Cook G.A., Park E.A. Peroxisome proliferator activated receptor alpha (PPARalpha) and PPAR gamma coactivator (PGC-1alpha) induce carnitine palmitoyltransferase IA (CPT-1A) via independent gene elements. Mol Cell Endocrinol. 2010;325:54–63. doi: 10.1016/j.mce.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan Y.J., Morimoto M., Thurman R.G., Bojes H.K., French S.W. Expression of the peroxisome proliferator-activated receptor gene is decreased in experimental alcoholic liver disease. Life Sci. 1995;56:307–317. doi: 10.1016/0024-3205(94)00953-8. [DOI] [PubMed] [Google Scholar]
- 55.Zeng T., Zhang C.L., Song F.Y., Zhao X.L., Xie K.Q. CMZ reversed chronic ethanol-induced disturbance of PPAR-alpha possibly by suppressing oxidative stress and PGC-1alpha acetylation, and activating the MAPK and GSK3beta pathway. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan Y.Y., Cai Y., Li J., Yuan Q., French B., Gonzalez F.J., French S. Regulation of peroxisome proliferator activated receptor alpha-mediated pathways in alcohol fed cytochrome P450 2E1 deficient mice. Hepatol Res. 2001;19:117–130. doi: 10.1016/s1386-6346(00)00089-9. [DOI] [PubMed] [Google Scholar]
- 57.Puigserver P., Adelmant G., Wu Z., Fan M., Xu J., O'Malley B., Spiegelman B.M. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 58.Azahri N.S., Di Bartolo B.A., Khachigian L.M., Kavurma M.M. Sp1, acetylated histone-3 and p300 regulate TRAIL transcription: mechanisms of PDGF-BB-mediated VSMC proliferation and migration. J Cell Biochem. 2012;113:2597–2606. doi: 10.1002/jcb.24135. [DOI] [PubMed] [Google Scholar]
- 59.Ghare S.S., Joshi-Barve S., Moghe A., Patil M., Barker D.F., Gobejishvili L., Brock G.N., Cave M., McClain C.J., Barve S.S. Coordinated histone H3 methylation and acetylation regulate physiologic and pathologic fas ligand gene expression in human CD4+ T cells. J Immunol. 2014;193:412–421. doi: 10.4049/jimmunol.1400055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furlan-Magaril M., Rincon-Arano H., Recillas-Targa F. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol Biol. 2009;543:253–266. doi: 10.1007/978-1-60327-015-1_17. [DOI] [PubMed] [Google Scholar]